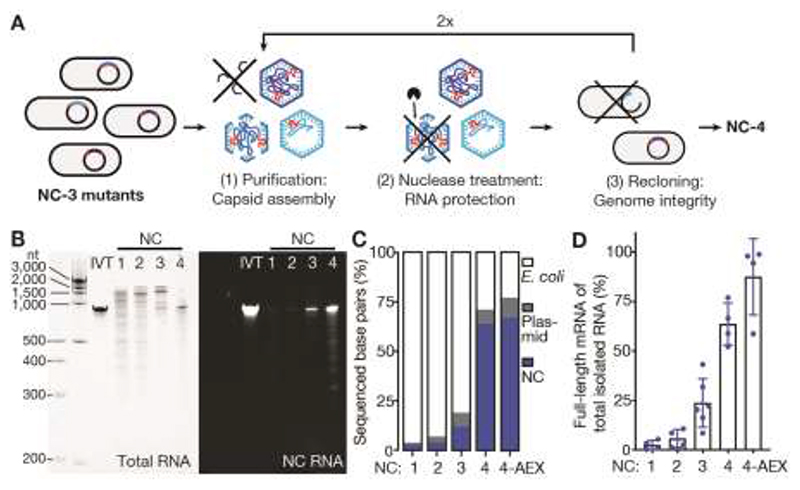
Fig. 1. NC-4 packages its genome with high selectivity.
(A) Laboratory evolution: a library of NC-3 mutants generated by error-prone PCR was expressed in Escherichia coli and purified by affinity and size exclusion chromatography. This step recovers assembled capsids. The purified capsid library was then treated with nucleases to enrich for capsids that protect their RNA cargo. Finally, the RNA was extracted from capsids, reverse-transcribed, and re-cloned into the original expression vector. This step selects for capsids that contain full-length genomes. (B) Denaturing PAGE (5%) of NC-1 to NC-4 stained for total RNA with GelRed (left) and the fluorogenic dye DFHBI-1T (right), which selectively binds the broccoli aptamer present in the 5’ - and 3’-untranslated regions of the mRNA genome (NC RNA). IVT, in vitro-transcribed reference mRNA. (C) RNA identities and their relative abundance were determined by Oxford Nanopore Sequencing ( 32 ) for all four capsids, including anion-exchanged NC-4 (4-AEX), and assigned to three main categories: bacterial RNA (E. coli), nucleocapsid mRNA (NC), and RNA originating from other plasmid-associated genes (plasmid). The encapsulated E. coli genes are primarily rRNA (Fig. S3). (D) The fraction of total extracted RNA corresponding to the full-length mRNA genome was determined by real-time quantitative PCR (mean of at least two biological replicates, each measured in two separate laboratories, error bars represent the standard deviation of the mean).