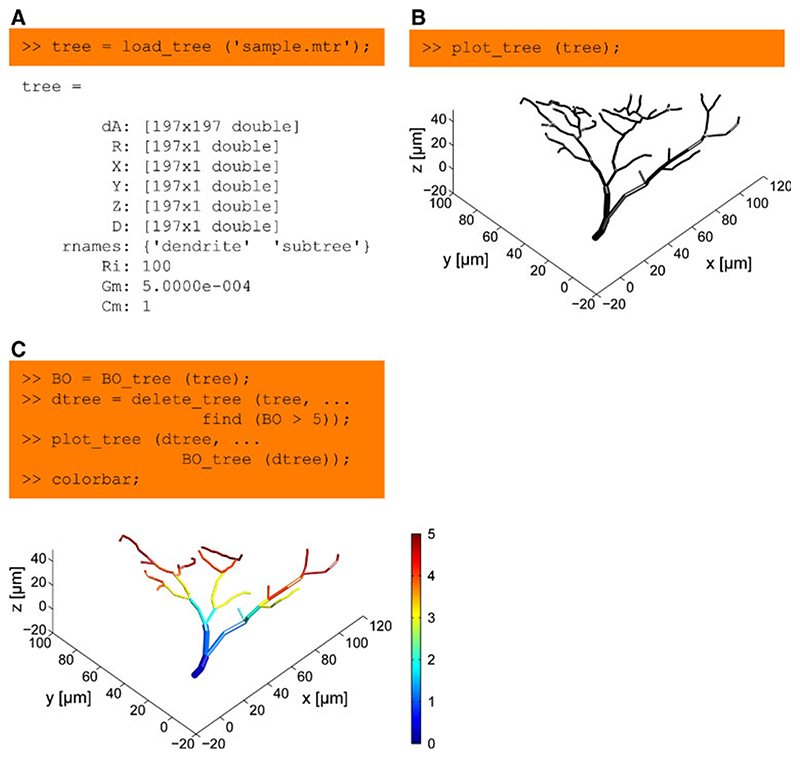
Fig. 1. Basic handling of a dendritic tree using the TREES toolbox.
a A tree is represented as a structure with: a directed adjacency matrix dA attributing a parent to each node in the tree; a set of vertical vectors, such as X, Y and Z coordinates and diameter values D, each assigning a value to every node in the tree; an array rnames containing sub-region names; arbitrary optional quantities such as the global electrotonic properties Ri, Gm and Cm, specific axial resistance, membrane conductance and membrane capacitance respectively. b Most functions in the TREES toolbox take a tree as their first argument. This is the case for example for the function plot_tree, which allows a 3D interactive visualization of the sample tree loaded in (a). c Statistics and tree edit functions are generally simple and easy to use. Here the branch order (BO) values of the tree are obtained using BO_tree and all nodes with BO larger than 5 are deleted using delete_tree. The remaining tree is visualized and BO values are mapped in pseudo-colour onto the tree using the optional second argument of the function plot_tree. Axis labels were added for clarity