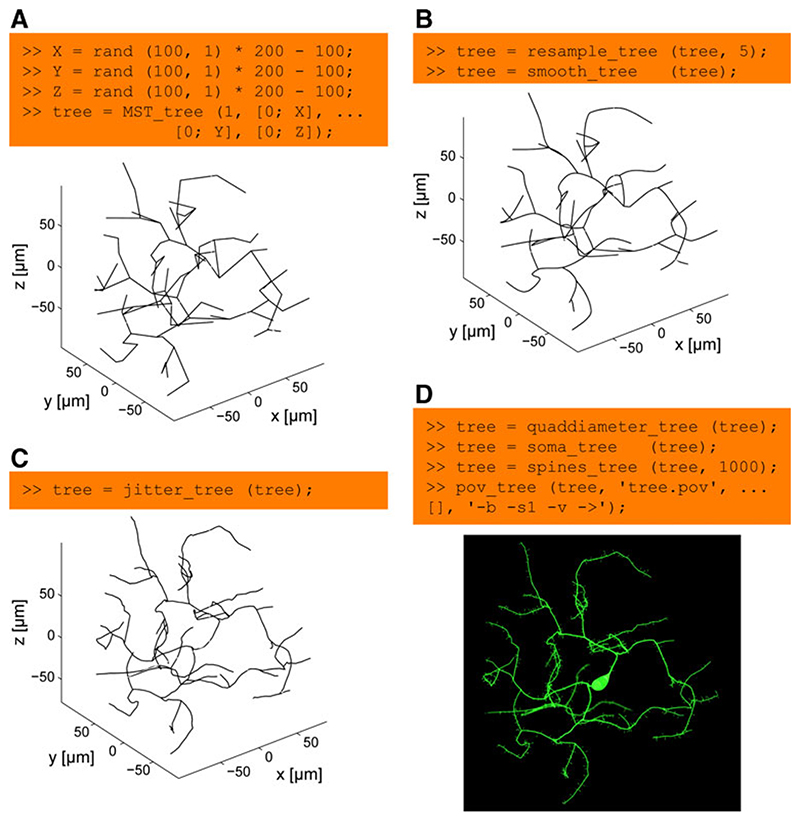
Fig. 3. The modular process of generating synthetic neuronal trees.
a 100 points at random locations in the range between -100 and 100 μm are connected to a root in (0, 0, 0) using the extended minimum spanning tree algorithm implemented in MST_tree. This step is quick: run time of 50 ms on a standard computer. b The resulting tree is resampled using resample_tree to produce densely distributed nodes along the tree at 5 μm intervals. This allows the node locations to be altered to smooth the tree structure with smooth_tree (run time: 200 ms). c And, subsequently, to add biologically realistic spatial noise with jitter_tree. This step can take a bit longer (run time: 1 s in this case). d In a last step, a diameter taper is mapped onto the tree structure using quaddiameter_tree, a soma-like diameter protuberance is mapped onto the root of the tree using soma_tree and spines_tree distributes spine structures randomly along the tree. Finally, the morphology is sent in one command (pov_tree) to the POV-Ray renderer for visualization. Note that this entire process is modular and each step can be replaced by alternative algorithms. The artificial neuron generated in this figure does not reproduce a particular real counterpart since the target points were simply chosen randomly within a 200×200×200 μm cube. Axis labels were added for clarity