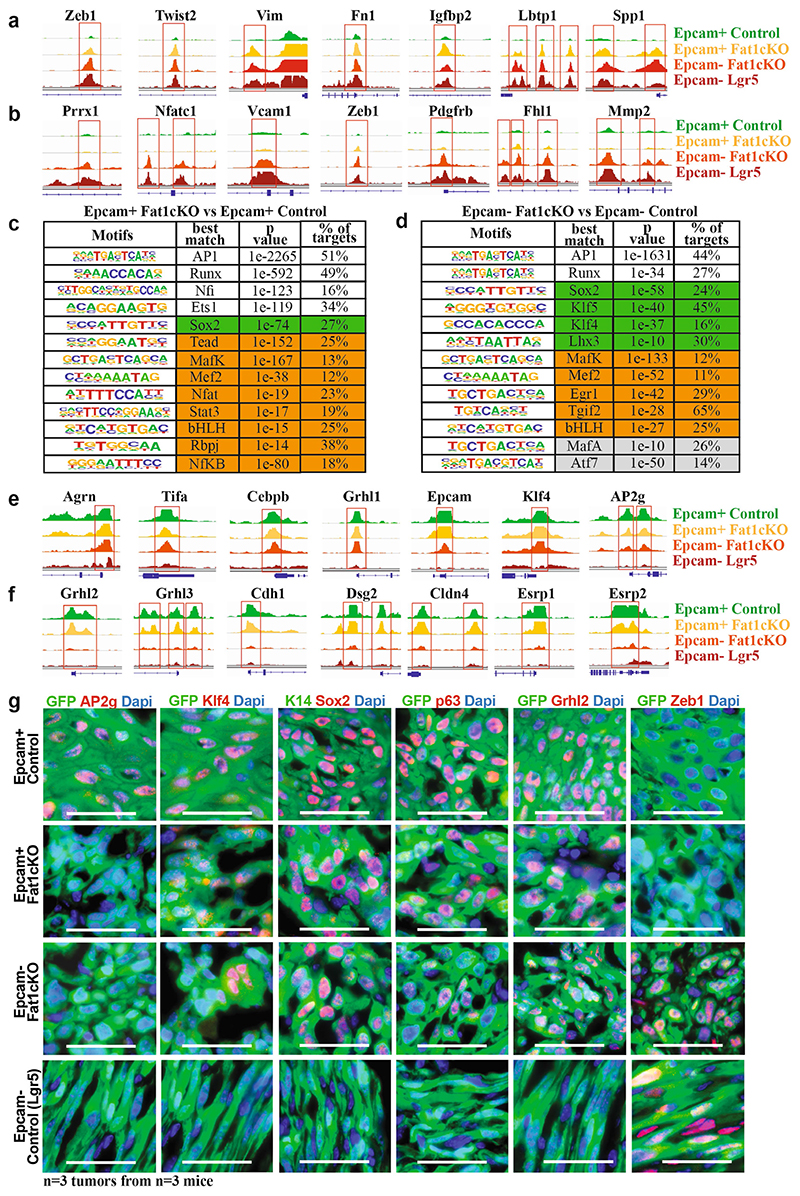
Extended Data Fig. 6. EPCAM+ Fat1-cKO tumour cells are epigenetically primed to undergo EMT, whereas EPCAM− Fat1-cKO sustain the expression of epithelial program.
a, ATAC-seq profiles of the chromatin regulatory regions of mesenchymal genes closed in control EPCAM+ tumour cells and opened in EPCAM+ Fat1-cKO tumour cells, showing epigenetic priming of EPCAM+ Fat1-cKO tumour cells to undergo EMT. b, ATAC-seq profiles of the chromatin regulatory regions of mesenchymal genes with open chromatin regions only in EMT EPCAM− tumour cells. c, Transcription factor motifs enriched in the ATAC-seq peaks upregulated between the EPCAM+ Fat1-cKO and EPCAM+ control tumour cells as determined by Homer. Cumulative hypergeometric distributions. White boxes show core transcription factors; boxes highlighted in green show epithelial transcription factors; and boxes highlighted in orange show EMT transcription factors. d, Transcription factor motifs enriched in the ATAC-seq peaks that are upregulated between the EPCAM− Fat1-cKO and EPCAM− control tumour cells as determined by Homer analysis. Cumulative hypergeometric distributions. White boxes show core transcription factors; boxes highlighted in green show epithelial transcription factors; boxes highlighted in orange show EMT transcription factors; and boxes highlighted in grey show other transcription factors. e, ATAC-seq of the chromatin regulatory regions of epithelial genes with open chromatin regions in EPCAM− Fat1-cKO tumour cells as compared to EPCAM− tumour cells from LGR5-derived SCCs, showing the sustained opening of epithelial enhancers in EPCAM− Fat1-cKO tumour cells. f, ATAC-seq of the chromatin regulatory regions of epithelial genes that are closed upon EMT, irrespective of Fat1 deletion. g, Immunostaining for GFP and AP2G, KLF4, SOX2, p63, GRHL2 or ZEB1 in EPCAM+ and EPCAM− control and Fat1-cKO DMBA/TPA skin SCCs. Scale bar, 50 μm.