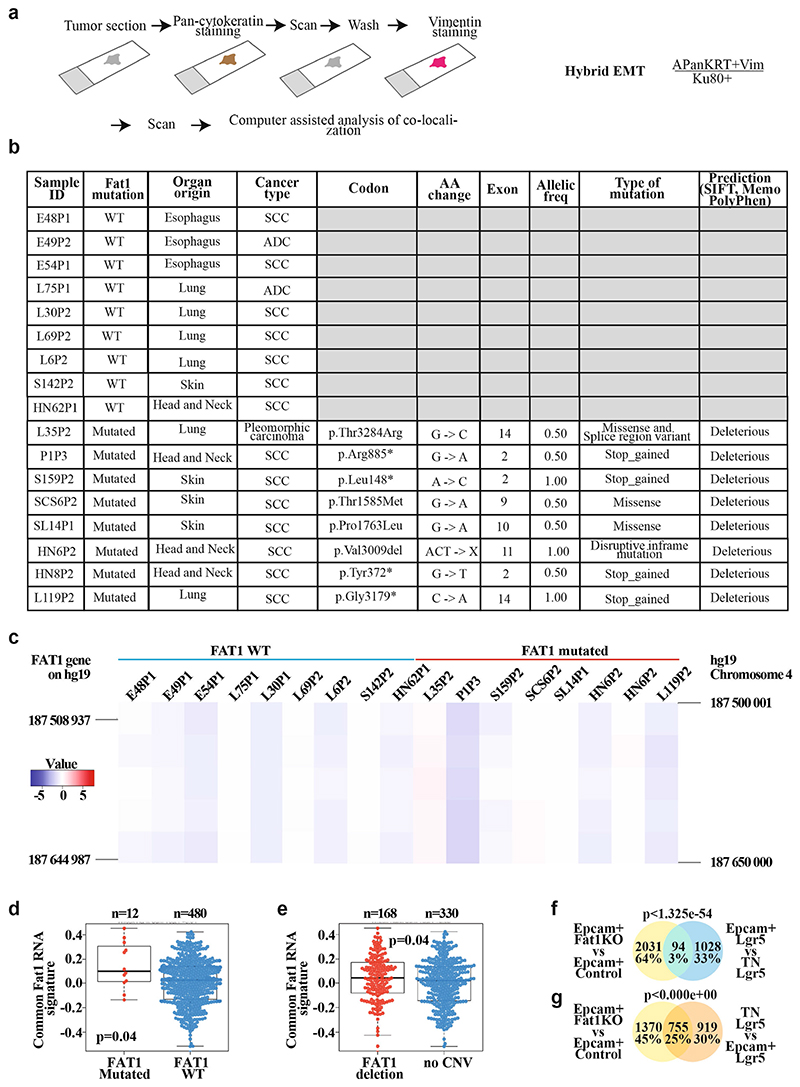
Extended Data Fig. 5. Mutations in FAT1 promotes hybrid EMT state in human cancers.
a, Schematic representing the method of analysing the co-expression of pan-cytokeratin and vimentin using immunohistochemistry of patient-derived xenografts that present (or not) mutations in FAT1, and the definition of hybrid EMT score. b, Table summarizing the samples of patient-derived xenografts on which whole-exome sequencing was performed, and detailed information on the mutations: codon, amino acid change, the exon containing the mutation, the allelic frequency, the type of mutations and the bioinformatic prediction of the effect of the mutation on the function of the protein using three bioinformatic algorithms (SIFT, Memo and PolyPhen). c, Heat map showing the copy number variation profile of FAT1 genomic region in the patient-derived xenograft samples included in the analysis of hybrid EMT score. The colour code corresponds to the quantified copy number and the genomic coordinate (reference genome hg19) of bin set for quantification. The FAT1 gene is marked on each vertical edge. d, e, Box plot showing the distribution of the common mRNA signature (mouse skin and lung Fat1-cKO SCCs and human FAT1-knockout SCC cell line) compared to FAT1 mutation status in human lung SCC (TCGA database; for the analysis, only high-impact mutations in >20% of variant allele frequency were considered) (d) and FAT1 copy number variation status in human lung SCC (TCGA database) (e). Boundaries of the box indicate the first and third quartiles of the FAT1 RNA signature value. The bold horizontal line indicates the median and the two external horizontal lines shows the minimum and maximum values. The dots represent all data points. Differences between the two groups are assessed using a two-sided Wilcoxon rank-sum test. f, g, Venn diagram of the genes upregulated in the EPCAM+ Fat1-cKO skin SCC and upregulated in LGR5 EPCAM+ versus triple-negative hybrid EMT tumour cells (f) or in triple-negative versus EPCAM+ cells (g). Two-sided hypergeometric test.