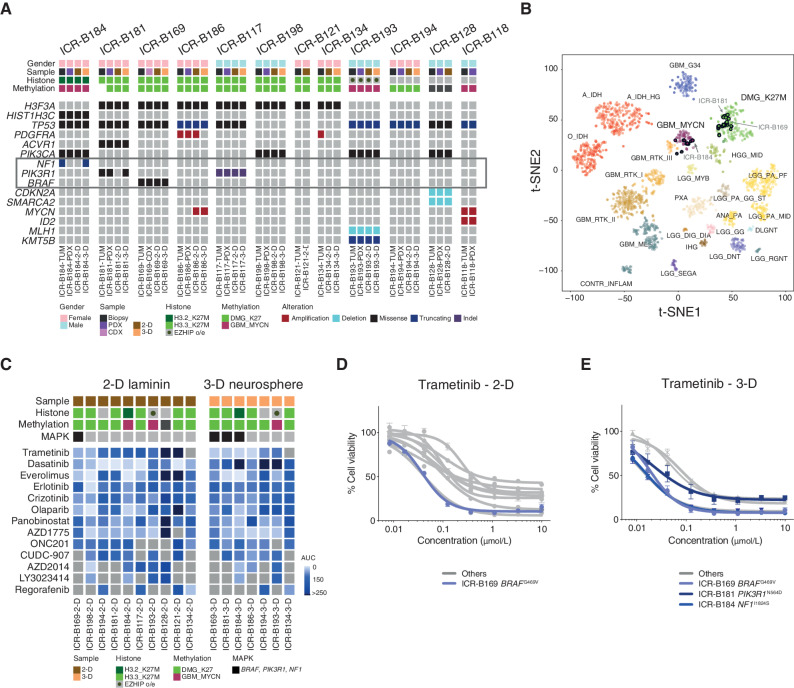
Figure 1.
In vitro sensitivity to trametinib in patient-derived DIPG models. A, Oncoprint representation of an integrated annotation of single-nucleotide variants, DNA copy number changes, and structural variants for patient-derived models and tumor biopsy specimens. Samples are arranged in columns with genes labeled along rows. Clinicopathologic and molecular annotations are provided as bars according to the included key. B, The t-statistic–based stochastic neighbor embedding (t-SNE) projection of a combined methylation data set comprising the in vitro models (circled) plus a reference set of glioma subtypes (n = 1,766). The first two projections are plotted on the x- and y-axes, with samples represented by dots colored by subtype as labeled on the figure. C, Drug sensitivities in the mini-screens carried out on cells grown under 2-D and 3-D conditions, visualized by heat map of normalized AUC values. Clinicopathologic and molecular annotations are provided as bars according to the included key. D, Dose–response curves for the MEK inhibitor trametinib tested against patient-derived models in vitro grown in 2-D. E, Dose–response curves for the MEK inhibitor trametinib tested against patient-derived models in vitro grown in 3-D. Cells harboring MAPK pathway alterations are highlighted in blue. Concentration of compound is plotted on a log scale (x-axis) against cell viability (y-axis). Means plus standard errors are plotted from at least n = 3 experiments.