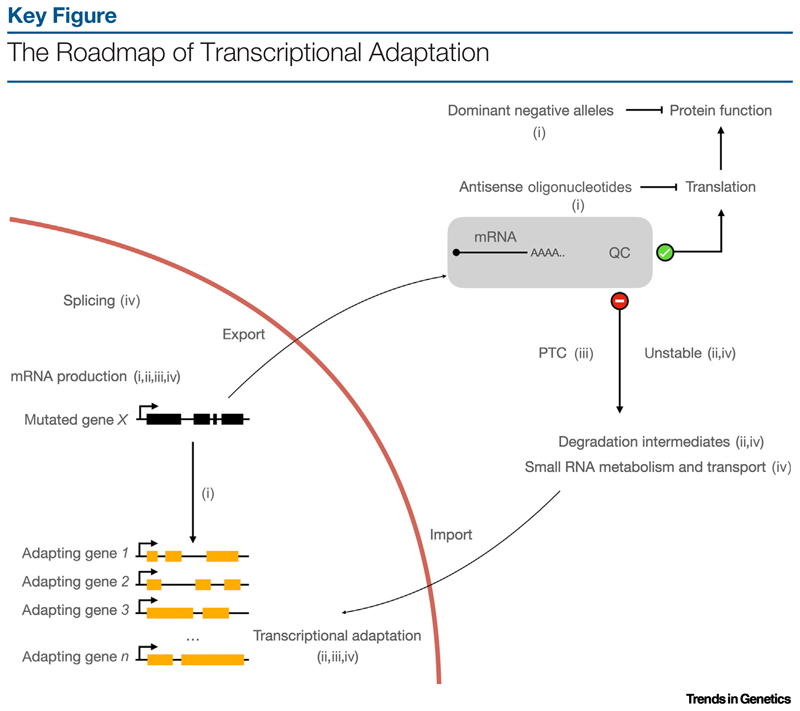
Figure 1.
Data from key studies {(i), Rossi et al. [39]; (ii), El-Brolosy et al. [48]; (iii), Ma et al. [49]; (iv), Serobyan et al. [52]} have helped build a basic framework for transcriptional adaptation: a frameshift mutation in gene X can activate the transcription of similar genes in trans [39,48,49,52]. This activation is not dependent on loss of protein activity as transcriptional [39,48,49,52] or translational [39] inhibition of gene X, or dominant negative alleles [39], do not trigger this response. The mRNA quality control (QC) mechanism of the cell determines whether an mRNA is used for protein production or is recycled [48,49,52]. If not used for translation, mRNAs can also enter the transcriptional adaptation pathway, either repurposed as long noncoding RNAs [49] or by contributing small degradation intermediates [48,52]. Different processes have been implicated in transcriptional adaptation, either upstream or downstream of the QC step [48,49,52]. Better understanding of the crosstalk between these processes will help explain different transcriptional adaptation responses and allow modulation of this pathway. Abbreviation: PTC, premature termination codon.