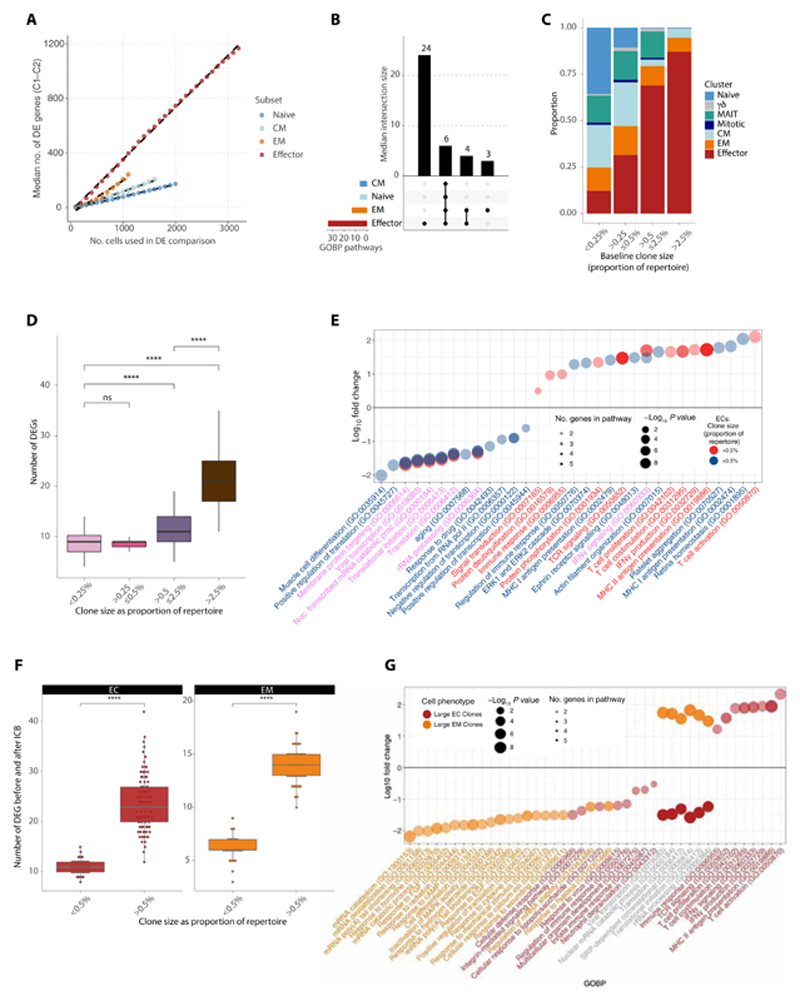
Fig. 3. Subset-specific and clone size-dependent transcriptome changes upon ICB.
(A) Median number of DE genes across 21 days of ICB in conventional T cell subsets. Each subset was subsampled to the indicated number of cells at d0 and d21 and tested for DE genes across 100 bootstraps. At each n value for number of cells, there was a significant difference across the subsets in the number of DE genes (P < 0.0001, Wilcoxon rank sum test). (B) Upset overlap plot of GOBP up-regulated in each subset after treatment (subsample of n = 1100 cells), based on the DE genes per bootstrap in (A). The median number of pathways in each intersecting set was calculated over the 100 bootstraps. (C) Clonal size bins at d0 filled by proportion of each phenotypic cluster. (D) Number of DE genes before and after treatment per clonal size bin. Each clonal size bin was subsampled to a constant size and the analysis bootstrapped 100x. (E) GOBP analysis demonstrating pathways up- and down-regulated in large versus small ECs before and after treatment. Large ECs are colored in bright red and small ECs in blue. Pathways where the text is in purple represent those that are shared between both groups. (F) Number of DE genes before and after treatment for large versus small ECs (red) and EMs (orange). Each cluster was subsampled to a constant size and the analysis bootstrapped 100x. (G) GOBP analysis demonstrating pathways up- and down-regulated in large ECs (red) versus large EMs (orange) before and after treatment. Pathways where the text is in gray represent those that are shared between both groups. For all analysis comparing numbers of DE genes, P values were determined by two-sided Wilcoxon rank sum test. ns, not significant.