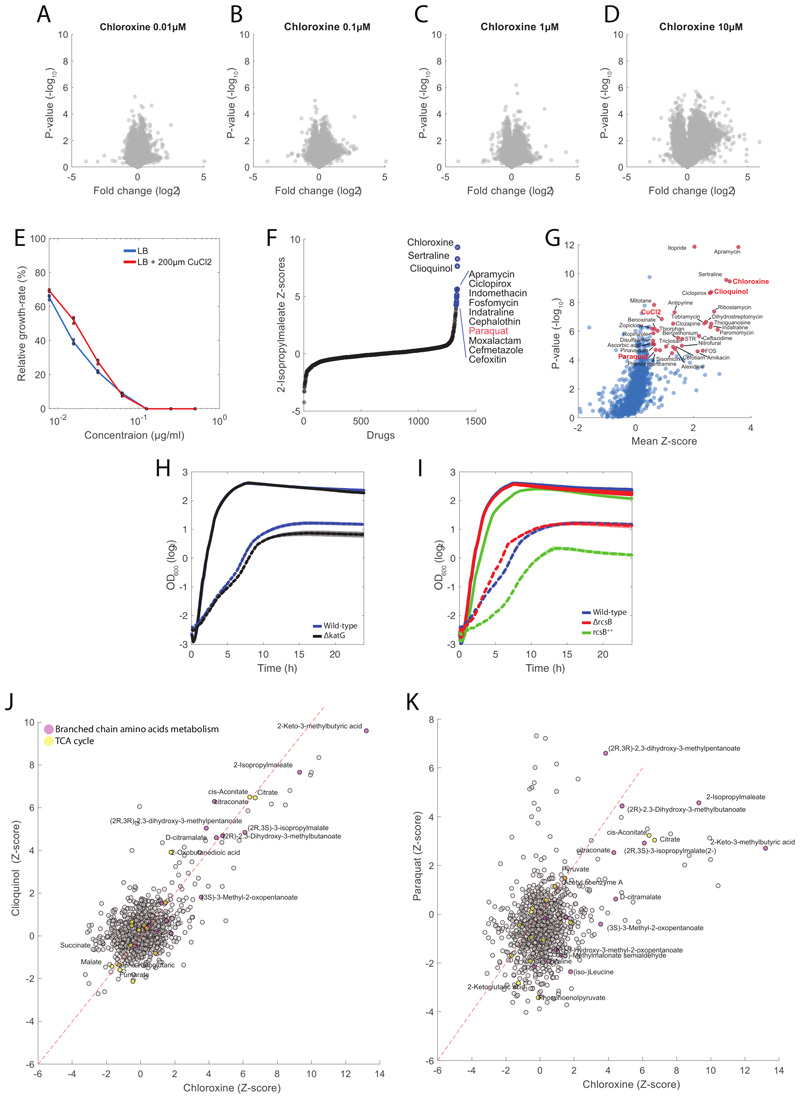
Extended Data Fig. 7. Chloroxine induce copper toxicity.
A-B-C-D) Results from limited proteolysis analysis. Each dot in the volcano plot represents the relative difference in peptide abundance between the treated and untreated proteome extracts. None of the 13797 detected peptides exhibit significant (pvalue<7.2480e-07, Bonferroni corrected) conformational changes between untreated and whole proteome extracts treated with 4 different concentrations of chloroxine (Table S6). Pvalues are estimated by twosided t-test analysis. E) Relative growth rates at different concentrations of chloramphenicol with respect to the untreated condition. Wild type E. coli was grown in LB medium with and without 200 μM of CuCl2. Growth inhibitory activity for different chloramphenicol concentrations were measured in triplicates. Errorbars are mean ± standard deviation across three biological replicates. F) 2-Isopropylmaleate z-score levels across the 1342 tested compounds. G) Average Z-score across metabolites in the branched chain amino acids biosynthesis pathway as defined in KEGG. Significance (i.e. pvalue estimated by permutation test) of enrichment analysis is testing for each compound an average increase of metabolite levels. Out of the 1342 tested compounds (i.e. Prestwick library + antibiotics), only 35 (less than 3%) induced significant changes in the levels of BCAA intermediates (pvalue≤5e-4 Bonferroni corrected and mean Z-score≥0.5). In addition to chloroxine, clioquinol, CuCl2 and paraquat, we observed that several of the 35 compounds inhibit protein synthesis (e.g. apramacyn, tobramycin, ribostamacyn...). This is likely reflecting the indirect accumulation of amino acids, including BCAA intermediates, as a results of the inhibition of protein synethesis4,18. H) Growth of wild-type E.coli (blue) and ΔkatG (black) without and with (dashed lines) 7.8μM of chloroxine. H) Growth of wild-type E.coli (blue), ΔrcsB (black) and rcsB++ (expression of rcsB is under the control of an IPTG-inducible promoter, here we used 1mM of IPTG) without and with (dashed lines) 7.8μM of chloroxine. J) Scatter plot of metabolic changes induced by chloroxine vs clioquinol. K) Scatter plot of metabolic changes induced by chloroxine vs paraquat. Some of the largest metabolic changes induced by paraquat involved metabolic intermediates of branched chain amino acids.