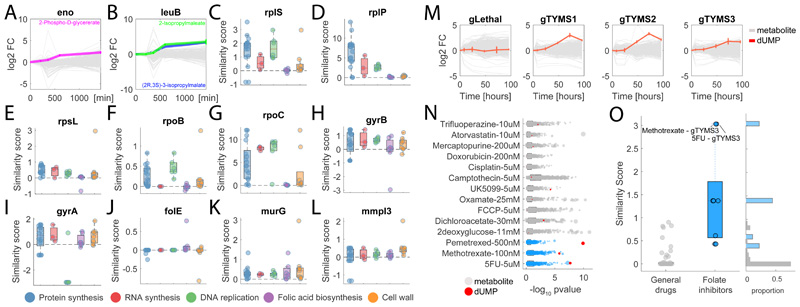
Figure 3. Metabolic profiling of chemical and genetic perturbations in M. smegmatis and A549 cancer cells.
A-B) Time-course profiles of changes in intracellular metabolite abundances after CRISPR-mediated knockdown of enolase (eno) and 3-isopropylmalate dehydrogenase (leuB) in M. smegmatis. Reported data are average over 3 biological replicates. C-L) Each dot represents the metabolic similarity (i.e. similarity scores) between genetic and chemical perturbations in M. smegmatis. Antimicrobials are grouped in 5 major antibiotic classes and the distribution of similarities with gene knockdowns metabolic profiles are illustrated by box plots. The tops and bottoms of each box are the 25th and 75th percentiles, respectively, while the red line in the middle of each box is the samples median. The lines extending above and below each box are the whiskers. Whiskers extend from the ends of the boxes delimited by the interquartile to the largest and smallest observations. M) Time-course profiles of changes in intracellular metabolite abundances in A549 lung cancer cells after CRISPR-mediated knockout of thymidylate synthase (TYMS), using three individual guide RNAs targeting different gene exons and a separate guide RNA targeting multiple genomic regions (gLethal) is used to induce general growth inhibition (Extended Data Figure 5). Changes in dUMP are highlighted in red. Reported data are average ± standard deviation over 3 biological replicates. N) Metabolic changes induced by 14 drugs with diverse targets and MoAs. Each dot corresponds to a putatively annotated metabolite, the log2 fold-change is reflected in the dot size, and statistical significance (p-value estimated by by two-tailed t-test analysis) on the x-axis. Drugs targeting enzymes in folate metabolism are highlighted in blue, changes in dUMP abundance in red. O) Boxplot of metabolic similarity (i.e. similarity scores) between genetic and chemical perturbations in A549 human lung cancer cells. Similarities between genetic and chemical interference with TYMS (blue) are compared to all remaining pairwise similarities (grey). The tops and bottoms of each box are the 25th and 75th percentiles, respectively. The lines extending above and below each box are the whiskers. Whiskers extend from the ends of the boxes delimited by the interquartile to the largest and smallest observations.