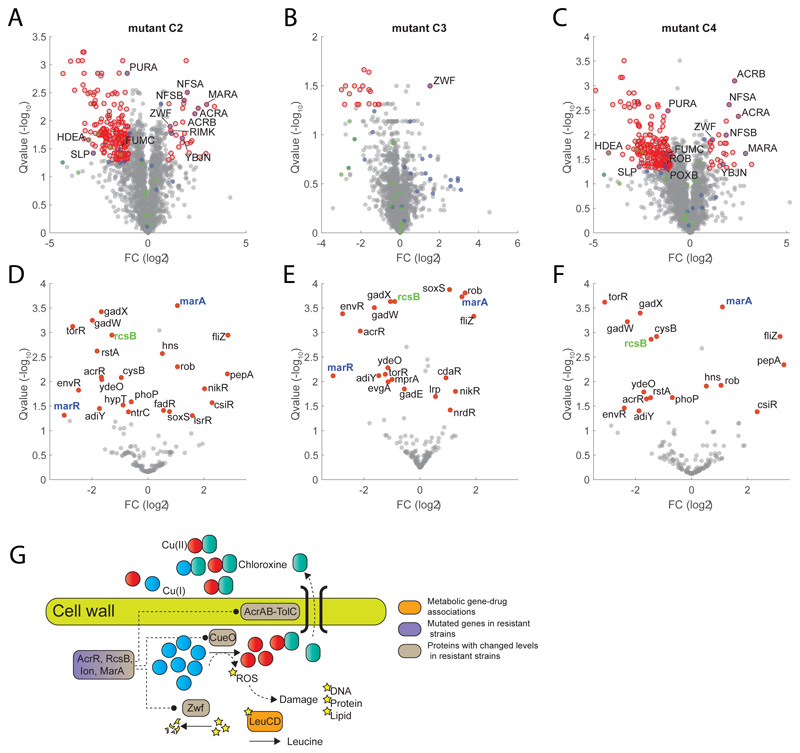
Figure 6. Chloroxine resistance.
A-B-C) Volcano plot of proteome changes in three chloroxine resistant strains (Supplementary Dataset 7) with respect to wild-type E. coli (Supplementary Dataset 8). Significant changes (qvalue≤0.05 and |FC|≥1) are highlighted in red. Proteins that are transcriptionally regulated by marA or rcsB are highlighted in blue and green, respectively. Reported data are average over three biological replicates. Pvalues estimated from two-sided ttest analysis are corrected for multiple tests by q-value estimation49. D-E-F) Transcription Factor differential activity profiles. For each TF, we reported average fold-changes of regulated proteins and significance of the difference with respect to wild-type (qvalue) estimated using a permutation test (see Materials and Methods). TFs with significant changes in activity are highlighted in red (qvalue≤0.05). G) Schematics of metabolome derived gene-chloroxine associations and their relation to genetic and proteome changes in chloroxine resistant strains.