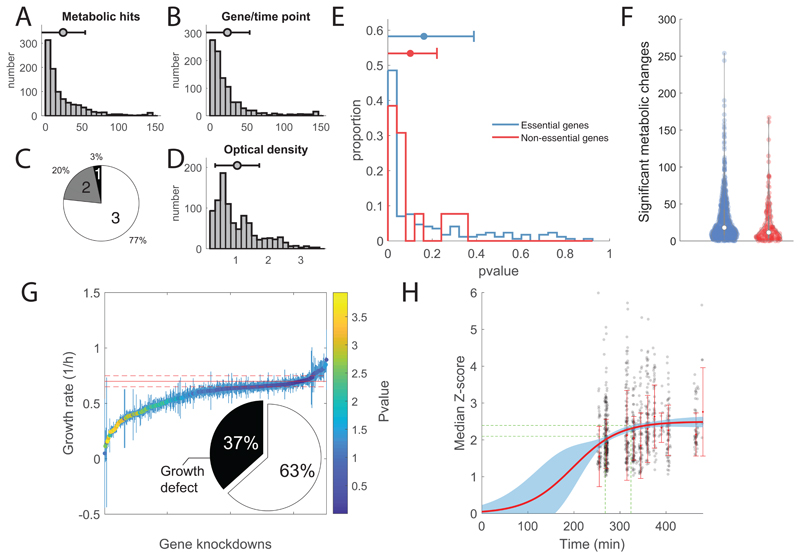
Extended Data Fig. 1. Metabolome profiles of CRISPRi mutant library.
A) Distribution of significant metabolic changes per gene knockdowns/time points. On average, 24 metabolites underwent significant changes (absolute Z-score ≥ 1 and p value ≤ 1e–5) per mutant and time point, and most of the metabolites (95%) exhibited a significant response in at least one gene knockdown. B) Distribution of significant changes for each metabolite across knockdowns/time points. C) Relative distribution of collected time points per gene knockdown. D) Distribution of Optical Densities (OD600) at sampling. E) Distribution of locality scores for essential (blue) and non-essential knockdowns (red). F) Distribution of the number of significant affected metabolites by essential (blue) and non-essential (red) gene knockdowns in each time point. G) Estimates of growth rates of CRISPRi mutants grown in M9 glucose + 1mM IPTG. Each dot corresponds to mean ± standard deviation (SD) across three biological replicates. Average Wild-type growth rate ± SD are represented by solid and dashed red lines, respectively. Each dot is color-coded according to the significance in growth rate difference with respect to wild-type (i.e. t-test analysis). Pie chart reports on the fraction of knockdown strains growing significantly slower than wild type (pvalue≤0.01). H) Median absolute Z-score of significantly affected metabolites (|Z-score| ≥ 1 and p value ≤ 1e–5) in each mutant/time point. Median values for each timepoint across all mutant strains are used to fit a sigmoidal curve (red curve). The blue shaded area identifies the confidence region (95% confidence interval). Green dashed lines report the 95% confidence interval for the time necessary to reach 90% of the maximum estimated changes (i.e. plateau).