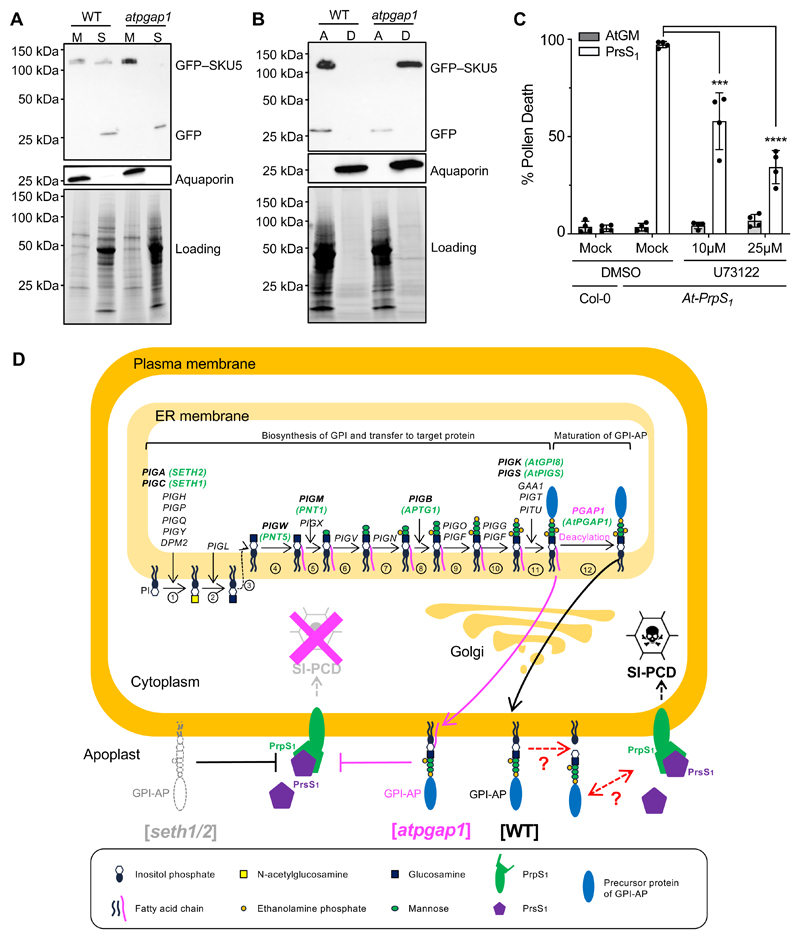
Figure 6. Evidence and a model for a requirement of GPI-APs at the plasma membrane for SI.
(A) Membrane (M) and soluble (S) fractions from WT and atpgap1 (atpgap1-c1) plants were extracted and localization of GFP–SKU5 (~120 kDa) proteins were analyzed on western blots. In the WT, GFP–SKU5 was detected in both fractions, but in the atpgap1 mutant, GFP–SKU5 was only detected in the membrane fraction, consistent with defective cleavage. Aquaporin (~26 kDa) is shown as a control for membrane extraction; loading control shows signals from a PROTEAN TGX Stain-Free Precast gel (Bio-Rad).
(B) Whole proteins were extracted using 2% Triton X-114 extraction buffer, partitioned into aqueous (A) and detergent (D) phases. Western blot analysis showed that GFP–SKU5 (~120 kDa) was primarily enriched in the aqueous phase in the WT, but in the atpgap1 mutant it was primarily detected in the detergent phase, revealing that mutation of AtPGAP1 resulted in an increase of GFP–SKU5 hydrophobicity. Aquaporin (~26 kDa) is shown as a control for membrane extraction; loading control shows signals from a PROTEAN TGX Stain-Free Precast gel (Bio-Rad).
(C) Col-0 and At-PrpS1 pollen grains were pretreated with the PLC inhibitor, U73122, or solvent (Mock) before subject to SI induction (PrsS1) or control treatment (AtGM) in vitro and samples were co-stained with FDA and PI 6h after treatment. Four independent experiments were carried out; 100-200 pollen grains were counted for each treatment. At-PrpS1 pollen treated with recombinant PrsS1 and mock solvent had high levels of death; treatment of At-PrpS1 pollen with recombinant PrsS1 and U73122 resulted in significant lower levels of death. This suggests that cleavage of GPI-APs by PLC is required for mediating SI-induced death of pollen.
(D) GPI-anchoring is a post-translational modification involving several phases which have been established in mammals and yeast. First, biosynthesis of the GPI involves a series of events [steps 1-10]. Once the core GPI is assembled [step 10], the target precursor protein (blue sphere) is transferred to the GPI by a GPI transamidase complex [step 11]. The nascent immature GPI-AP then undergoes remodeling; the first of these is deacylation [step 12], involving PGAP1, which we identified in this study (indicated in pink). After this, several other PGAP genes (not shown) mediate further modification during the transport from ER to the Golgi and then to the plasma membrane. The genes involved in GPI biosynthesis identified in humans are indicated in black; genes identified in plants are indicated in green. Knockouts of genes early in the biosynthetic pathway in mammals result in lack of expression of GPI-APs at the plasma membrane; in plants homozygous knockouts of the orthologs of PIG-C and PIG-A, SETH1 and SETH2, are lethal. We show in this study that mutation of SETH1 or SETH2 in At-PrpS1 pollen alleviates SI-induced pollen death. By analogy to animal systems, it is likely that the seth1/2 mutants lack GPI-APs at the plasma membrane (grey outline). Our data implicate GPI-APs at the plasma membrane are required for the SI response, as the normal SI-PCD response is prevented in their absence. After generation of the nascent GPI-APs, the acyl chain linked to inositol is removed by the GPI-deacylase, PGAP1 [step 12]. In the atpgap1/hld1 mutant, this step does not occur. As a consequence, the GPI-APs retain their inositol-linked acyl chain (indicated in pink), giving a 3-footed configuration responsible for retaining the GPI-AP in the plasma membrane in mammalian cells. Here we provide evidence that this is also the case in plants. We show that knockout of PGAP1 completely prevents SI, resulting in full fertility. This demonstrates that inositol deacylation, required for maturation of GPI-APs, is critical for the SI response. We propose that during an incompatible SI induction when the ligand PrsS interacts with plasma-membrane localized PrpS, this S-specific interaction may require interaction (direct or indirect) with (unknown) GPI-APs. Cartoon adapted from 9.
See also Figure S5.