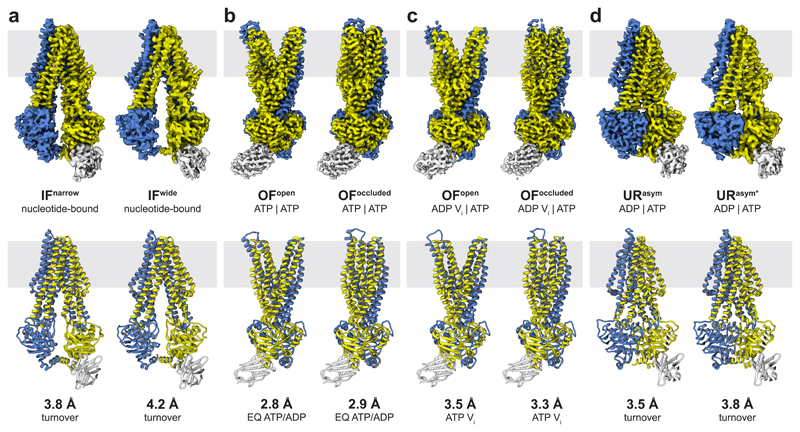
Figure 1. The conformational space of TmrAB.
a, Two distinct IF conformations from TmrABturnover. In IFwide, the intracellular gate has opened by 4.4 Å, allowing access for bulky substrates. Wide and narrow IF conformations were also found in the control datasets and in absence of nucleotides and/or substrate. Densities for nucleotides were observed in all IF conformations in TmrABturnover and TmrAEQBATP-ADP datasets, but not in TmrABapo and TmrABC4F, as no nucleotides were added in the latter cases. b, In the ATP-bound state, the NBDs are tightly dimerized and the exporter exhibits either an OFopen or OFoccluded conformation. The same two OF conformations are observed in both TmrAEQBATP and TmrAEQBATP-ADP datasets (E523Q mutation in TmrA); therefore, only maps from TmrAEQBATP-ADP are shown here due to the higher resolution. c, OFopen and OFoccluded conformations also dominate in the vanadate-trapped state. d, The asymmetric states from TmrABturnover resemble an OFoccluded conformation with a slightly separated NBD interface. In contrast to the OFoccluded conformations of TmrAEQBATP-ADP and TmrABATP-Vi, the intracellular gate is opened by 1.5 Å and 3.0 Å in URasym and URasym*, respectively, while the extracellular gate is tightly sealed. The top and bottom rows show experimental maps and deposited models, respectively. OF densities and models in b and c are rotated by 90° with respect to panel a and d to illustrate the opening of the extracellular gate. TmrA is blue, TmrB yellow, and the nanobody light grey. All structures and morphs between conformers are highlighted in the Supplementary Video 1 and 2.