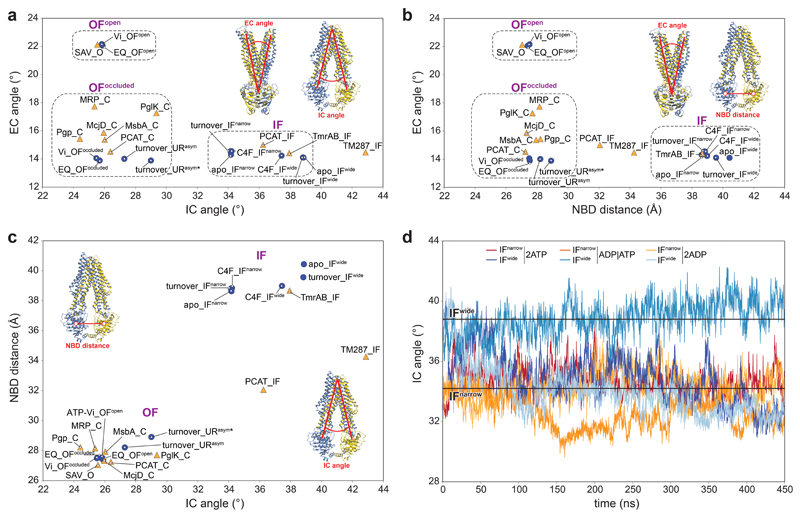
Extended Data Figure 7. Conformation space of type I ABC exporters.
a-c, Conformation space of the EM structures from this study and previously published ABC tranporter structures is shown based on three parameters: the intracellular gate angle (IC angle), the extracellular gate angle (EC angle), and the distance between two nucleotide-binding domains (NBD distance). The EM structures, obtained in this study, are shown as blue circles, and previously published structures as orange triangles. The clustering of structures in three major conformations (IF, OFopen, and OFoccluded) is highlighted by dashed rectangles in a and b. The EM structures, obtained in this study, cover all main conformations. The new URasym conformations have closest resemblance to OFoccluded. The IC angle is defined as the angle between two vectors: vector 1 between the center of mass of the whole extracellular part of the TMD region and the center of mass of the intracellular parts of TM1TmrA, TM2TmrA, TM3TmrA, TM6TmrA, TM4TmrB, and TM5TmrB; and vector 2 between the center of mass of the whole extracellular part of the TMD region and the center of mass of the intracellular parts of TM1TmrB, TM2TmrB, TM3TmrB, TM6TmrB, TM4TmrA, and TM5TmrA. The EC angle is defined as the angle between two vectors: vector 3 between the centers of mass of the NBDs and the extracellular parts TM1TrmA, TM2TrmA, TM3TrmB, TM4TrmB, TM5TrmB, and TM6TrmB; and vector 4 between the center of mass of the NBDs and of the extracellular parts TM1TmrB, TM2TmrB, TM3TmrA, TM4TmrA, TM5TmrA, and TM6TmrA. The NBD distance is defined by the centers of mass of two NBDs (N-terminal loops and C-terminal helices were excluded). IF: IF conformation; O: OFopen conformation; C: OFoccluded conformation; Vi: TmrABATP-Vi; EQ: TmrAEQB. PDB IDs: PCAT: 4RY2 and 4S0F for the IF and OFoccluded conformation, respectively51; McjD: 4PL016; Pgp: 6C0V52; MsbA: 5TTP53; PglK: 5C7354; MRP: 6BHU55; Sav: 2ONJ18; TmrAB: 5MKK12; TM287 (TM287/288): 4Q4A56. d, Time-dependent IC angle in independent MD simulations of IFwide and IFnarrow with ATP/ATP, ADP/ATP, and ADP/ADP in the canonical/non-canonical site, respectively.