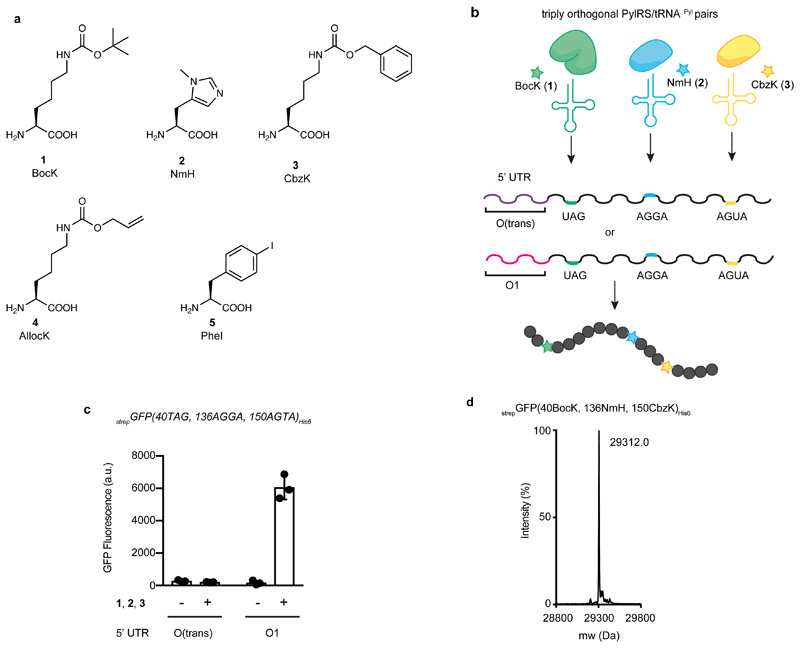
Figure 3. Efficient production of proteins containing three distinct ncAAs is enabled by new O-mRNAs.
a, Structures of the amino acids used in this work.N6-(tert-butoxycarbonyl)-L-lysine (BocK) 1; Nπ-methyl-L-histidine (NmH) 2; N6-((benzyloxy)carbonyl)-L-lysine (CbzK) 3; N6-((allyloxy)carbonyl)-L-lysine (AllocK) 4; (S)-2-amino-3-(4-iodophenyl)propanoic acid (PheI) 5. b, Engineered triply orthogonal pyrrolysyl-tRNA synthetase tRNA pairs for the incorporation of three distinct ncAAs using two different orthogonal messages. One message contains the O1-strepGFPHis6 5’UTR, generated by vol 1 of our algorithm, and the other message used the O-(trans) 5’UTR. c, Production of strepGFP(40BocK, 136NmH, 150CbzK)His6 from E. coli cells containing strepGFP(40TAG, 136 AGGA, 150 AGTA)His6 constructs with either the O(trans)- or O1-strepGFP-His6 5’UTRs. Cells also contained O-riboQ1 and the aaRS3/tRNA3 operons (encoding MmPylRS/MspetRNAPylCUA, MlumPylRS(NmH)/MinttRNAPyl-A17VC10UCCU and M1r26PylRS(CbzK)/MalvtRNAPyl-8UACU). ncAAs BocK 1, NmH 2, CbzK 3 were added to the cell. Each bar represents the mean of three biological replicates ± s.d. The individual data points are shown as dots. d, Results of positive electrospray TOF-MS of nickel-NTA purified strepGFP(40BocK, 136NmH, 150CbzK)His6 purified from cells. StrepGFP(40BocK, 136NmH, 150Cbz)His6 mass predicted: 29314.5, mass found: 29312.0. The experiment was performed three times with similar results.