Fig. 4. Leaf374 phyllosphere colonization is less affected when co-inoculated with Leaf245 EMS mutants that no longer secrete ASF05_00205 compared to the wild type.
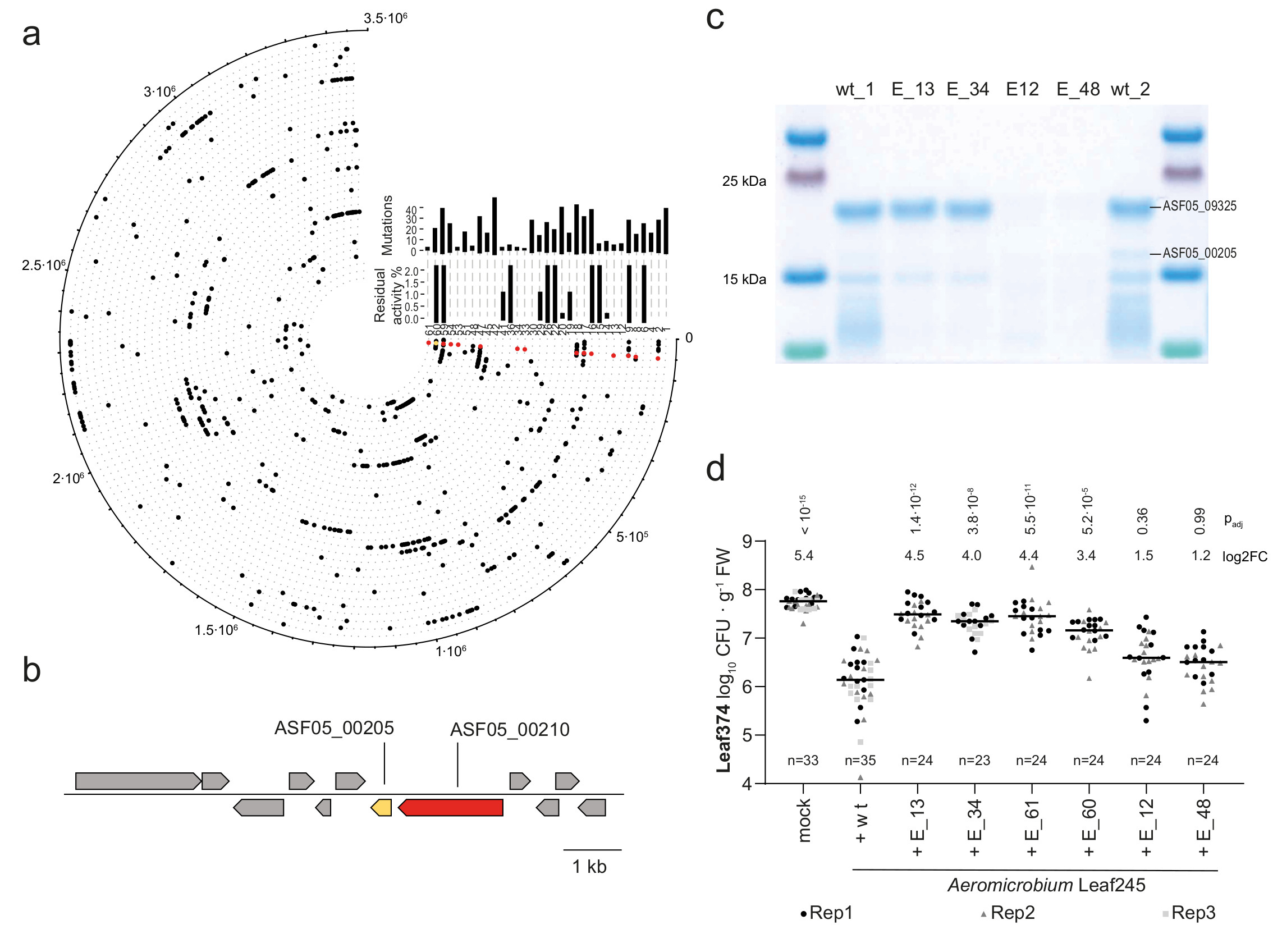
a) Graphic representation of mutations found in genome re-sequenced Leaf245 clones that no longer inhibit Leaf374 in vitro. Outermost circle indicates position on the Leaf245 genome and each circle with its corresponding number represents one clone. Total number of non-synonymous mutations and residual activity compared to the wild type strain are shown for each clone by the corresponding bar plots. Mutations in ASF05_00210 (red) and ASF05_00205 (yellow) are highlighted. b) Leaf245 genomic region comprising a gene pair encoding an identified regulator (red, ASF05_00210) and effector (yellow, ASF05_00205). c) SDS-PAGE protein bands observed in the active fraction of Leaf245 wild type and the corresponding fractions of selected Leaf245 EMS mutants. Protein bands with differing pattern between wild type and mutant strains are indicated with protein accession. A second biological replicate showed similar results. d) Leaf374 colonization of A. thaliana in mono association (mock) or in combination with Leaf245 wild-type or EMS mutants enumerated on MM maltose agar plates. The data of 2-3 biological replicates are shown. Each treatment was compared to Leaf374 colonization in combination with Leaf245 wild type, p-values (Kruskal-Wallis and post-hoc Dunn test, Bonferroni adjusted p) and log2 fold-changes are shown.
