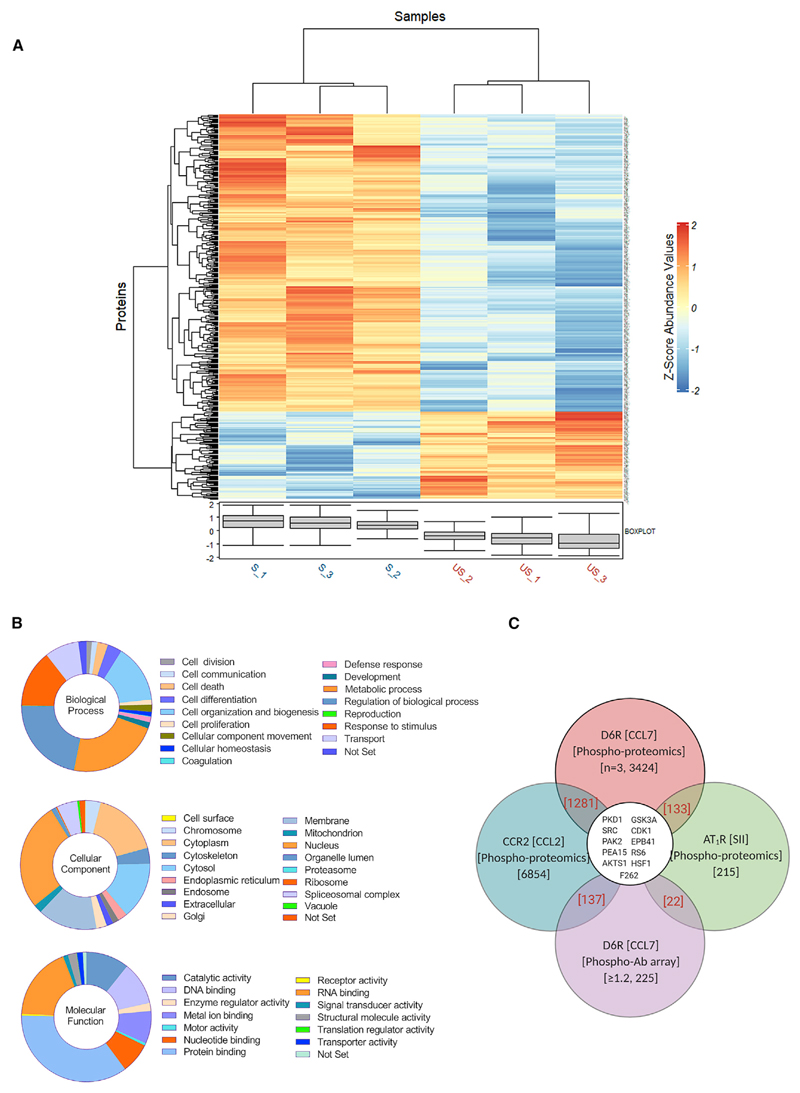
Figure 6. Phospho-proteomics reveals potential D6R signaling pathways.
(A) Heatmap depicting differentially phosphorylated proteins generated based on phospho-proteomics analysis on HEK293 cells expressing D6R with and without agonist stimulation. Three independent lysate samples prepared in parallel were used for trypsin digestion, enrichment of phospho-peptides using TiO2, and mass spectrometry (MS)-based identification of cellular proteins. S1–S3 indicate replicates of stimulated samples, while US1–US3 indicate replicates of unstimulated samples.
(B) Classification of cellular proteins that undergo phosphorylation/dephosphorylation upon D6R stimulation based on biological processes, molecular functions, and cellular localization reveal an extensive network of potential signaling pathways.
(C) Comparison of D6R phospho-proteomics data with phospho-antibody array and previously published phospho-proteomics study on CCR2 and AT1R reveals the activation of some common and multiple D6R-specific signaling proteins. Details of the common proteins are listed in Figure S6F. See also Figures S5–S7 and Tables S1 and S2.