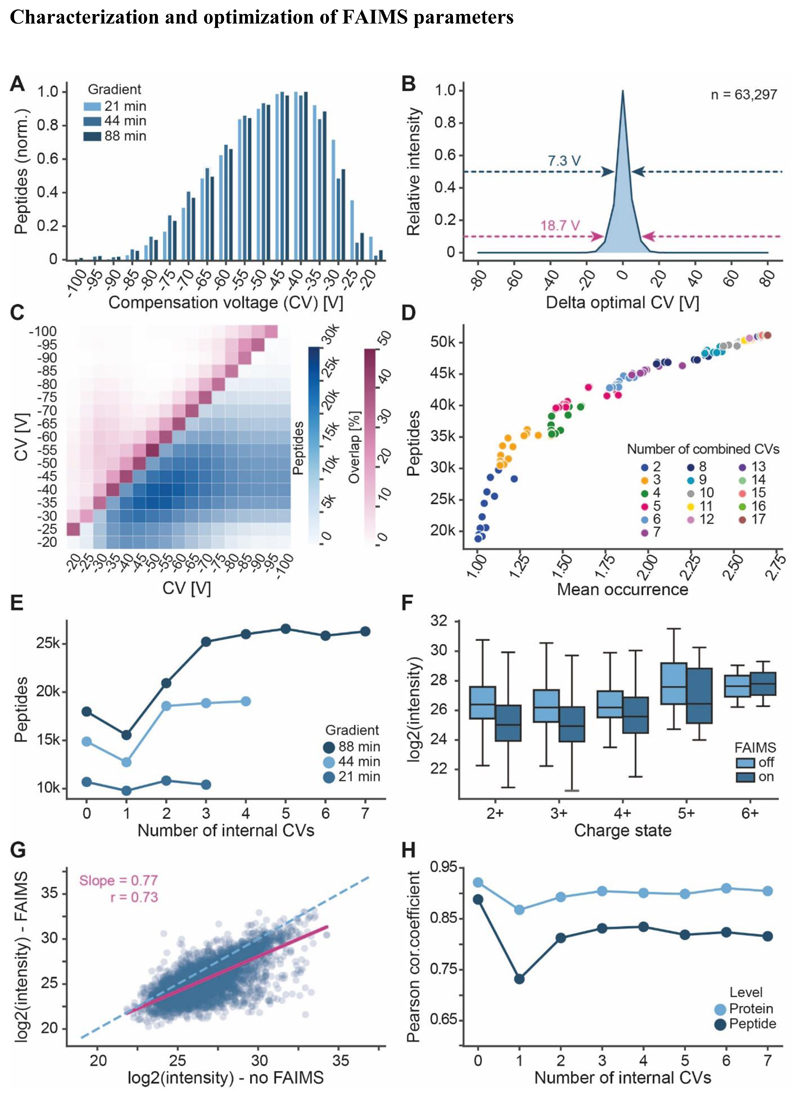
Figure 4. Characterization of FAIMS parameters.
(A) Distribution of peptide identification at different compensation voltages (CV) and gradient lengths. (B) Approximation of FAIMS resolution by analysing the intensities of peptides detected at different CVs relative to the intensity at their optimal CV (88 min gradient data shown). Arrows and values indicate the full width at half maximum (dark blue) or the width of the distribution at 10% maximal intensity (purple). (C) Absolute number of identified peptides (blue scale) and relative number of overlapping peptides (purple scale) for any two combined CVs (88 min gradient data shown). (D) In-silico analysis of the number of unique peptides that may be identified by combing between 2-17 CV values (based 88 min gradient data). (E) Number of actually identified peptides when combining several CVs within an nLC-MS/MS run of different gradient lengths. (F) Precursor ion intensity distributions of peptides detected in different charge states with or without FAIMS (CV=-45 V). (G) Scatterplot of peptide precursor ion intensities with or without FAIMS (CV=-45 V). The dashed line marks the diagonal and the purple line depicts the linear regression (r=Pearson correlation coefficient). (H) Correlation of peptide and protein group intensities of nLC-MS/MS runs applying the indicated number of internal CVs to the nLC-MS/MS run without FAIMS (For 0, another replicate without FAIMS was used).