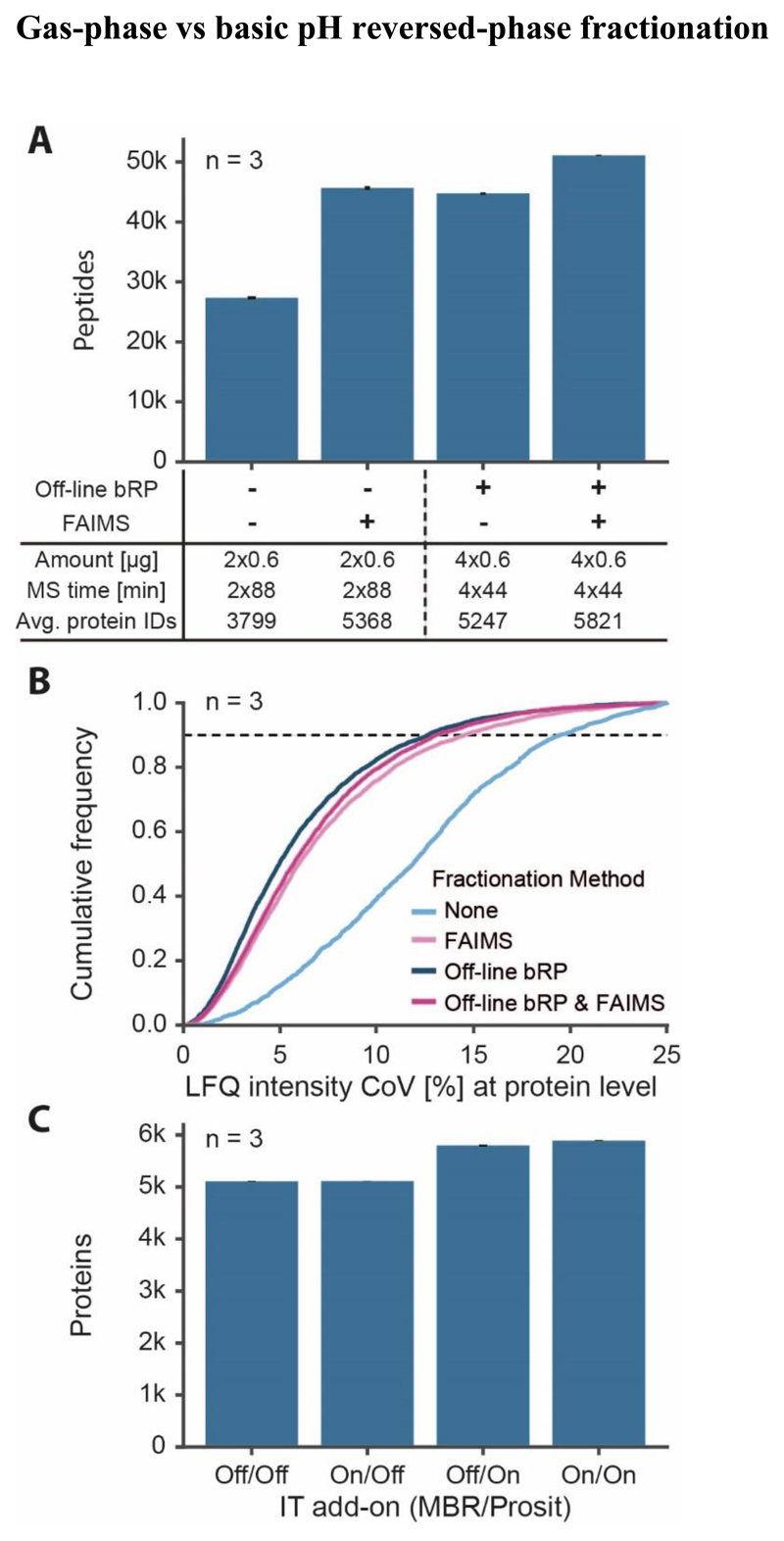
Figure 5. Comparison of proteomic depth obtained by FAIMS or off-line fractionation.
(A) Peptides identified (with SEM) with or without FAIMS and with or without basic pH reversed-phase fractionation (bRP; four fractions). Two sets of five CVs were combined for 88 min separations and 2 CVs were used for 44 min separations. (B) Empirical cumulative distribution function (ECDF) of the quantitative reproducibility described by the coefficient of variation (CoV) based on protein group intensities of three replicates (same data as for A). (C) Effect of using the ‘match between runs’ (MBR) feature of MaxQuant and/or Prosit rescoring on the number of identified proteins (with SEM, 88 min gradient with double injection using FAIMS with two sets of 5 CVs).