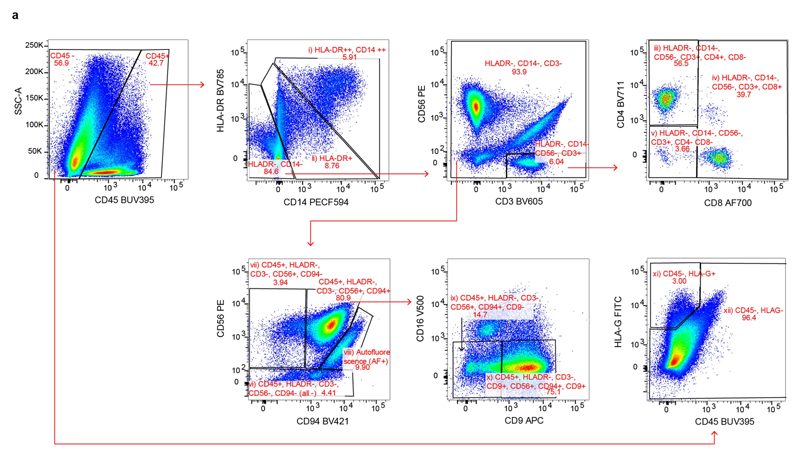
Extended Data Fig. 1. Gating strategy for Smart-seq2 data.
a, Gating strategy for a panel of 14 antibodies to analyse immune cells in decidual samples by Smart-seq2 (CD3, CD4, CD8, CD9, CD14, CD16, CD19, CD20, CD34, CD45, CD56, CD94, DAPI, HLA-DR and HLA-G). Cells isolated for Smart-seq2 data were gated on live; CD19- and CD20-negative, singlets and the following cell types were sorted: (i) CD45 CD14highHLA-DRhigh; (ii) CD45 HLA-DR ; (iii) CD45 HLA-DR CD56 CD3 CD4 CD8 ; (iv) CD45 HLA-DR CD56 CD3 CD8 ; (v) CD45 HLA-DR CD56 CD3 CD4 CD8 ; (vi) CD45 HLA-DR CD3 CD56 CD94 (labelled 'all -' on the figure); (vii) CD45 HLA-DR CD3 CD56 CD94 ; (viii) autofluorescence; (ix) CD45 HLA-DR CD3 CD56 CD94 CD9 ; (x) CD45 HLA-DR CD3 CD56 CD94 CD9 ; (xi) CD45 HLA-G ; (xii) CD45 HLA-G. Sample F9 is shown as an example. Cells from different gates were sorted in different plates: myeloid cells (gates (i) and (ii)); T cells (gates (iii), (iv) and (v)); natural killer cells (gates (vi), (vii), (viii), (ix) and (x)); CD45 (gates (xi) and (xii)). Antibody information is provided in Supplementary Table 10.