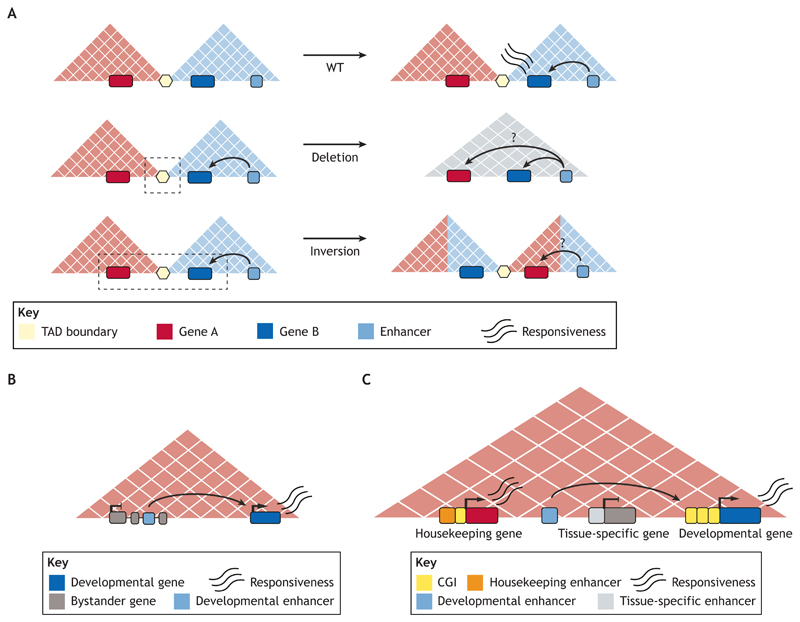
Fig. 1. The role of TADs in enhancer function and specificity.
(A) Enhancers and their target genes usually lie within the same TAD, enabling robust gene expression (top panel). However, structural variants can cause rearrangements of the genes and enhancers present within TADs. Deletions (middle panel) or inversions (lower panel) encompassing TAD boundaries can expose genes to new regulatory environments. Predicting the transcriptional effect of these rearrangements is not obvious, as genes do not always adopt enhancers located in the same TAD. (B) The majority of TADs contain several genes and enhancers. In these multi-gene TADs, enhancers associated with developmental genes can be found in the introns of neighboring genes with unrelated expression patterns (bystander genes). However, bystander genes do not respond to these enhancers, despite being within the same TAD, and are linked with their own regulatory elements. (C) The genetic diversity of promoters might explain the specificity of multi-gene TADs: housekeeping genes respond to housekeeping enhancers located within or close to the promoter regions; tissue-specific genes contain CpG-poor promoters and are responsive to proximal tissue-specific enhancers; while developmental genes contain promoters with large CpG island (CGI) clusters and respond to distal enhancers.