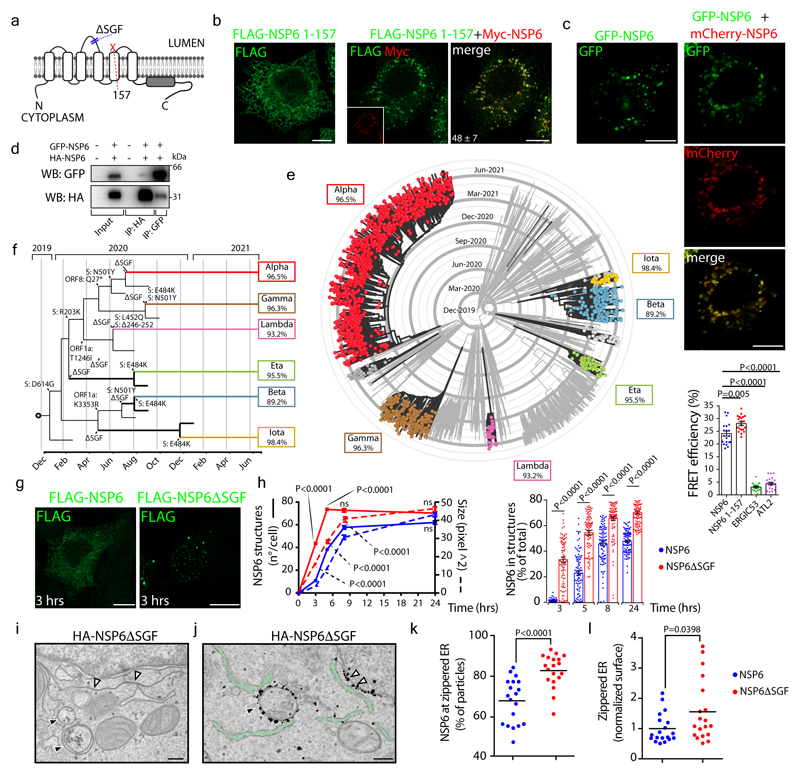
Figure 2. ER zippering requires NSP6 homodimerization and is more efficient with NSP6ΔSGF.
a, Predicted secondary structure of NSP6. The ΔSGF deletion and truncation site (at 157) are indicated. b, HeLa cells expressing Myc-NSP6 (inset) and/or FLAG-NSP6-1-157. The fraction of NSP6-1-157 associated with NSP6 structures is indicated. Mean ± SD, N=3, n=74. c, Cells expressing GFP-NSP6 alone or together with mCherry-NSP6. Graph, FRET measurements in cells co-expressing mCherry-NSP6 with the indicated GFP-tagged protein. Mean ± SD, N=3, n=20. d, Immunoprecipitation and Western blot (WB) from GFP-NSP6 and HA-NSP6 co-expressing cells, representative of four independent experiments. e, Radial layout of a phylogenetic tree of 3,508 SARS-CoV-2 genomes. VoCs are indicated and the percentage of each genome carrying ΔSGF is reported. Black branches highlight the appearance of the deletion. f, Mutations involved in branching and specificity of each VoC. Arrows, appearance of the ΔSGF and mutations in S protein. g, h, Time course analysis of stably-expressing FLAG-NSP6 or FLAG-NSP6ΔSGF cells induced with doxycycline. g, Fluoro-micrographs at 3 hrs. h, quantification of the structures shown in, g and Extended Data Fig. 5a, N=3, n=90. Left graph, number and size of NSP6-positive structures. Right graph, NSP6 in structures as percentage of total NSP6 in the cell. i, EM and j, IEM (anti-HA immunolabelling) of HA-NSP6ΔSGF-expressing HeLa cells. White and black arrowheads, linear and circular zippered-ER structures. Green, regular ER membranes. k, Morphometric analysis of IEM images. Quantification of gold particles at zippered ER (% of total ER-associated-particles). l, The surface area of zippered-ER normalized for the total number of gold particles. For k, l, N=3, n=19. Scale bars, b, c, g, 10 μm; i, j, 250 nm. Two-tailed Mann-Whitney test, c or unpaired two-tailed t-test, k, l, one-way ANOVA with Tukey’s post-hoc test, h. ns, not significant.