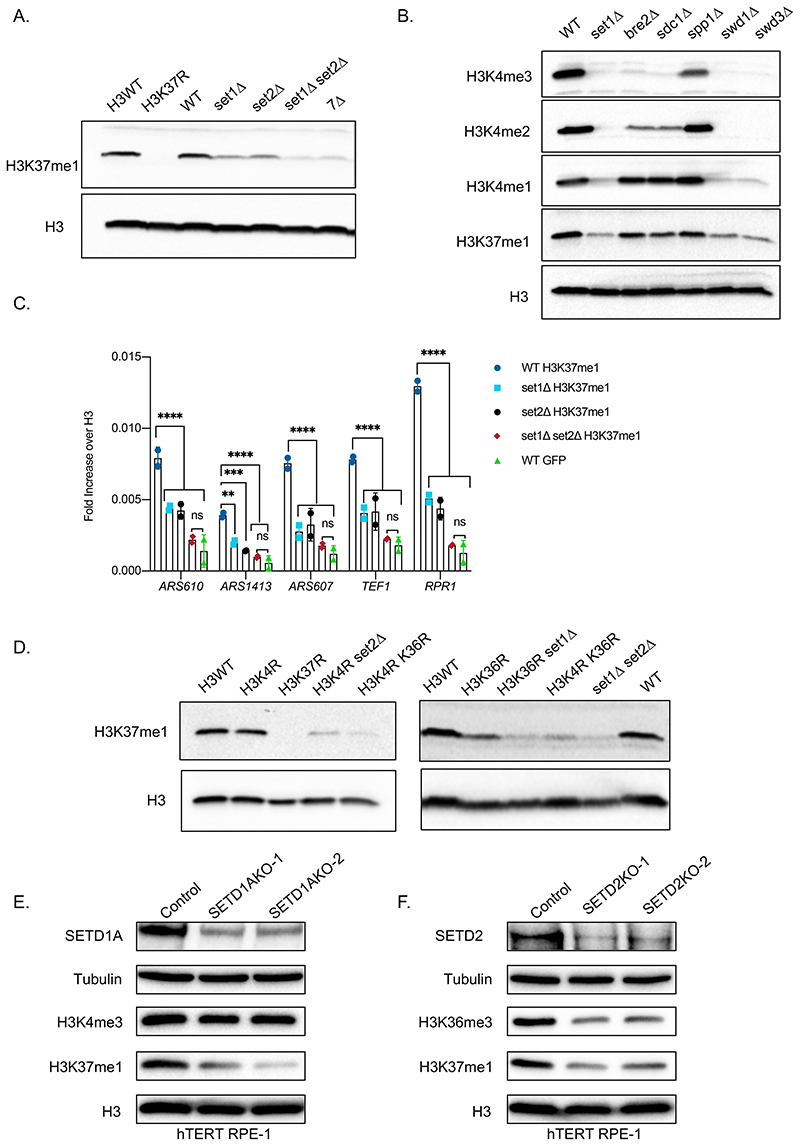
Figure 1. Set2 and Set1 (COMPASS) are necessary for H3K37me1 in vivo.
(A and B) Immunoblot analysis of total protein extracts from different yeast isogenic mutants as specified. Equal amounts of proteins were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 or anti-H3K4me1/me2/me3 antibodies and then re-probed with anti-H3 antibody. (C) ChIP qPCR experiments showing H3K37me1 levels at different genomic locations. Chromatin from isogenic strains was immunoprecipitated using anti-H3K37me1, anti-H3 and anti-GFP (negative control) antibodies. Statistical analysis was performed using Two-way ANOVA multiple comparisons and Tukey’s multiple comparison test (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments. (D) Immunoblot analysis of total protein extracts from different yeast isogenic strains as specified. Equal amounts of proteins were separated by SDS-PAGE in 16% acrylamide gels. Blots were probed with anti-H3K37me1 antibody and then re-probed with anti-H3 antibody as indicated. (E and F) Immunoblot analysis of total protein extracts from (E) SETD1A or (F) SETD2 CRISPR knockout cell pools. Human RPE1 cells stably expressing Cas9 were transfected with two guide RNAs targeting different exons of each gene or a non-targeting control. Total protein extracts were prepared 7 days after transfection, and proteins were separated by SDS-PAGE in 12% acrylamide gels or 3-8% Tris-Acetate gels.