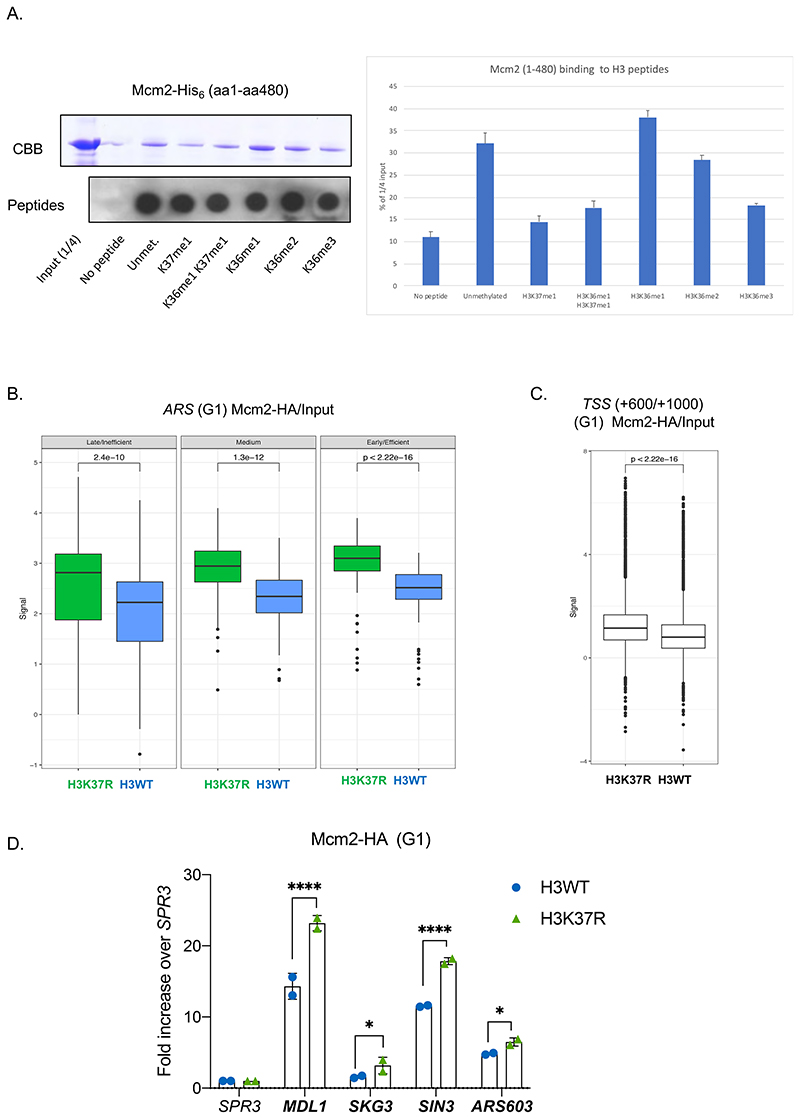
Figure 5. H3K37me1 regulates origin establishment.
(A) Left: Mcm2-His6 in vitro binding to biotinylated H3 peptides, modified as indicated. Input and peptide-bound Mcm2 were resolved by SDS-PAGE in 8% acrylamide and detected by Coomasie-Brilliant Blue (CBB) staining. 1/5 of each bead slurry was spotted onto PVDF membrane and biotin-peptides detected with anti-biotin antibody. Right: Average ImageJ quantification of assays shown in left and in Fig.S7A. Binding is represented as % of the signal corresponding to ¼ of the input. (B) Box plot of Mcm2-HA occupancy at early/efficient, medium and late/inefficient ARS +/- 1Kb. Blue: H3WT, green: H3K37R. (C) Box plot of Mcm2 occupancy at non-ARS (mean of the region from TSS+600 to TSS+1000) in H3WT and H3K37R mutant cells. The plots in (B) and (C) represent the average of the 3 independent cultures. (D) ChIP qPCR experiments showing Mcm2-HA levels at “H3K37R unique” replication sites. H3WT and H3K37R yeast cells expressing Mcm2-HA were arrested in G1, crosslinked and chromatin was immunoprecipitated (IP) with anti-HA antibody. The IP material was analyzed by qPCR with primers specific for each location. Data were normalized to the non-ARS region (SPR3). Statistical analysis was performed using Two-way ANOVA corrected for the comparisons using the Holm-Sidak method (Alpha: 0.05); * - P ≤ 0.05, ** - P ≤ 0.01, *** - P ≤ 0.001, **** - P ≤ 0.0001. Error bars represent the mean ± SD of 2 independent experiments.