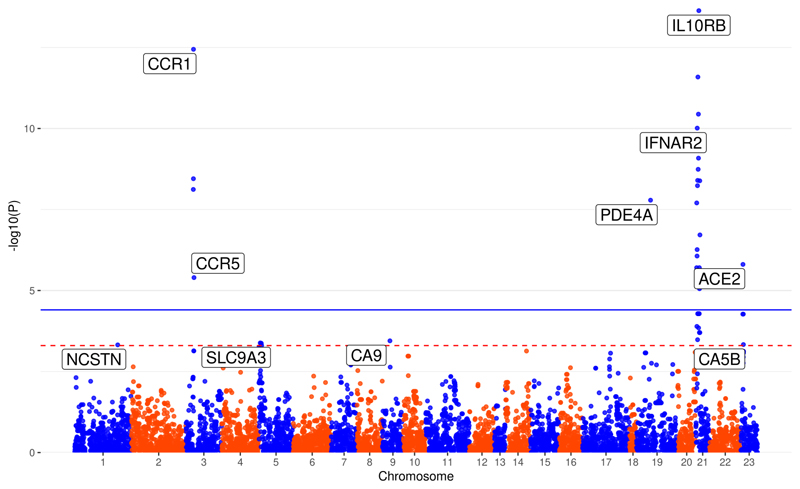
Figure 2. Manhattan plot of results from actionable druggable genome-wide Mendelian randomization analysis.
Mendelian randomization estimates were calculated using inverse-variance weighting and fixed effects for instruments that contained more than one variant, and Wald-ratio for instruments with one variant. Blue solid line indicates the P value threshold for significance (P<3.96×10-5, 0.05 Bonferroni-corrected for 1,263 actionable druggable genes) and red dashed line indicates a suggestive (P<5.00×10-4) threshold. Genes are labeled by their most significant MR association. For example, the results for IL10RB is most significant with cis-eQTL proposed instruments derived in skeletal muscle tissue (P=2.31x10-14), which is the point labeled. Results are plotted by the gene start position. All MR results with P value less than 5.00×10-4 used the GTEx cis-eQTLs as proposed instruments.