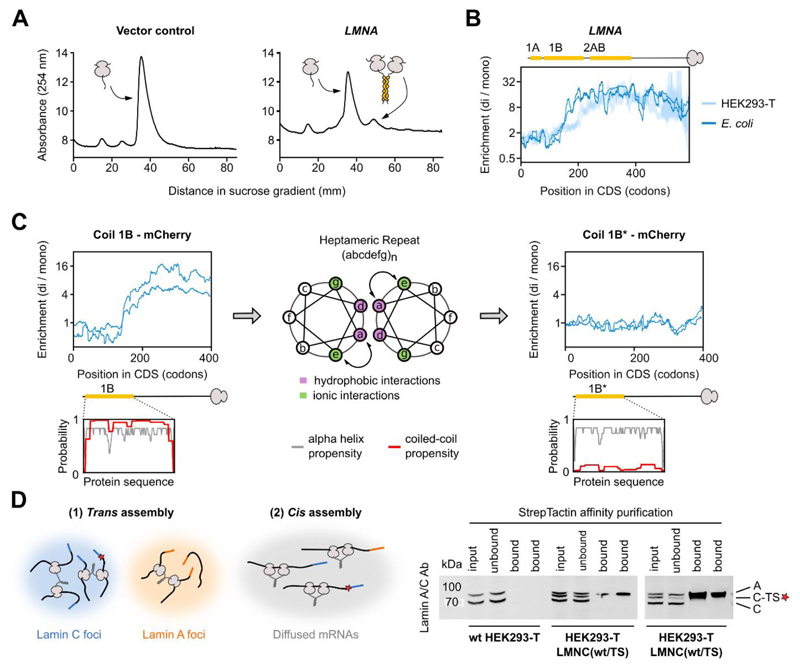
Fig. 5. Co-co assembly does not rely on eukaryote-specific factors and facilitates native biogenesis of lamin C homodimers.
A) Sucrose gradient sedimentation analysis of E. coli ribosomes from cells transformed with a control plasmid (left) or a plasmid enclosing human LMNA encoding lamin C (right), lacking the unstructured N-terminal head domain (33).
B) Disome over monosome enrichment profile of plasmid-encoded LMNA expressed in E. coli (dark blue, n = 2), and endogenously expressed LMNA in HEK293-T cells (light blue, n = 2). The ribosome-exposed coiled coil interfaces are indicated by yellow bars.
C) Disome over monosome enrichment profiles of LMNA encoding lamin coil 1B (left) or the a↔e swapped version of 1B (1B*, right) fused N-terminally to mCherry and expressed in E. coli (n = 2). The ribosome-exposed coiled coil interfaces are indicated by yellow bars. Helical wheel projection shows residue arrangements (a-g) of the heptad repeat (middle). Coiled coil (red) and alpha-helical (grey) probability predictions are shown for both wild type and mutant 1B (insets).
D) Hypothetical models of co-co assembly supporting isoform-specific homodimerization (left). A red star represents the TwinStrep tag (TS). Affinity purification of tagged lamin C (C-TS) from wild type or heterozygous LMNC(wt/TS) HEK293-T cells (bottom, technical replicates shown). Bands are labeled: A (lamin A), C (lamin C), C-TS (lamin C – TwinStrep).