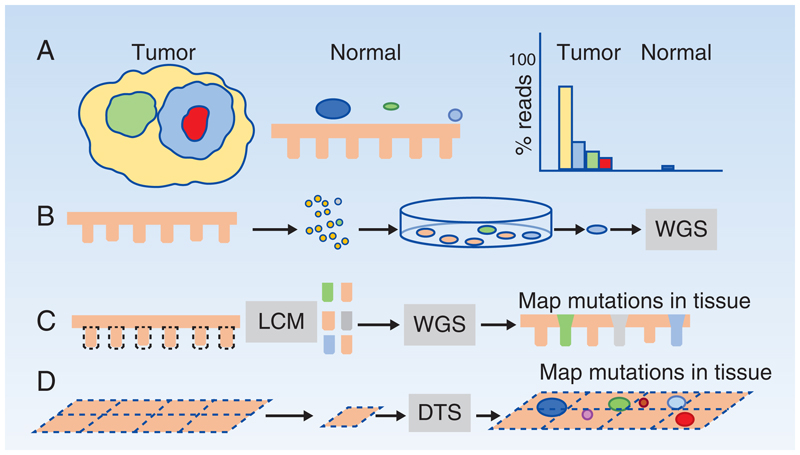
Figure 1. Detecting somatic mutants in normal epithelia.
A: Clonality and somatic mutant detection in tumors and normal epithelia. Tumors are clonal yellow) with sub-clonal mutations (green, blue and red). Normal epithelia harbor scattered somatic mutant clones (colored circles) in a wild type background). When sequenced at standard depth, the founder clone and common subclones in a tumor are readily identified, whereas only the largest mutant clones in normal tissue exceed the lower limit of detection.
B-D: Methods to detect somatic mutant clones in normal tissues
B: A single cell suspension is generated from normal epithelium, cultured at clonal density and individual colonies whole genome sequenced (WGS).
C: Laser capture microdissection (LCM) is used to isolate areas of tissue such as individual stem cell niches like the colonic crypt. Special protocols are used to perform WGS, whole exome or targeted sequencing and mutations can then be located within the tissue section from which they came.
D: Larger areas of epithelia can be dissected into a gridded array of typically 2mm2 samples, deep targeted sequencing (DTS) performed, and a statistical approach used to call rare somatic mutants in each sample, which can then be mapped (colored circles) within the sample grid.