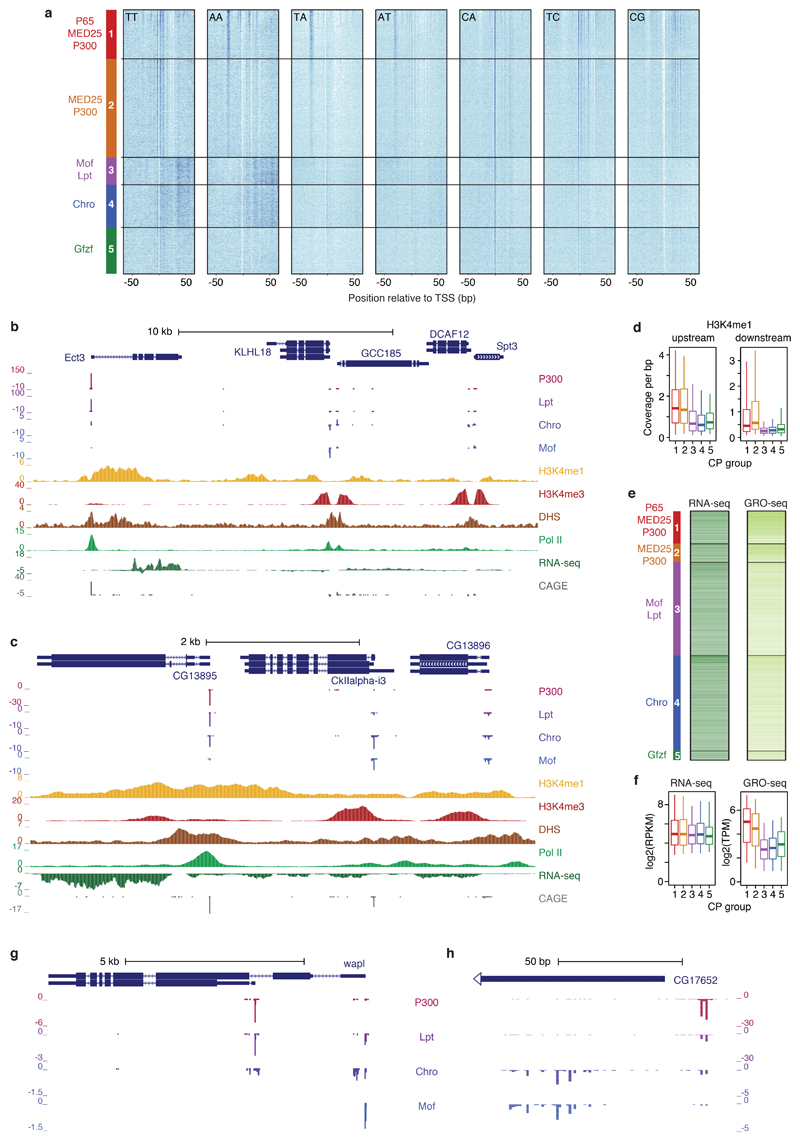
Extended Data Figure 9 |. Core promoters activated preferentially by distinct cofactors differ in their sequence and in endogenous chromatin features.
a, Occurrence of specific dinucleotides (see label in each heatmap) relative to TSSs for core promoters (CPs) of the five groups defined in Extended Data Fig. 6a. Within each group, CPs are sorted decreasingly by the COF-STAP-seq tag count of the respective strongest COFs (denoted on the left). Darker shade reflects higher density of the respective dinucleotides at specific positions. b, c, Examples of genomic loci with CPs active in S2 cells that are differentially activated by COFs in STAP-seq. All supporting data tracks are from S2 cells and re-analysed from previous publications (see Supplementary Table 1 for details and references). (b) CPs of KLHL18 and Spt3 (Group 3), and GCC185 and DCAF12 (Group 4), are preferentially activated by Mof and Chro, respectively, and have high levels of H3K4me3 downstream of their TSSs. In contrast, the CP of Ect3 (Group 1) is preferentially activated by P300 and has high levels of H3K4me1 both upstream and downstream of the TSS but almost no H3K4me3, although Ect3 is expressed and the CP is endogenously active in S2 cells. (c) CPs of CkIIalpha-i3 (Group 4) and CG13896 (Group 3) are preferentially activated by Chro and Mof, respectively, and both bear high levels of H3K4me3 and low levels of H3K4me1 downstream of the TSS. In contrast, the CP of CG13895 (Group 1) is preferentially activated by P300 and is marked by higher levels of H3K4me1, but lower levels of H3K4me3, although the gene is expressed in S2 cells. d, Average H3K4me1 ChIP-seq coverage in the 500 bp window upstream (left) and 500 bp window downstream (right) of the TSS for 5 groups of CPs active in S2 cells (as in Fig. 3b). n = 646, 363, 1842, 1885 and 179 CPs, for Groups 1 to 5, respectively. e, Heatmaps showing endogenous expression (as measured by RNA-seq [left] and GRO-seq [right]) of genes associated with CPs active in S2 cells from the 5 CP groups (RNA-seq and GRO-seq data from refs 44 and 45, respectively; see Supplementary Table 1 for details and references). Within each group, CPs are sorted decreasingly by STAP-seq of the respective strongest COFs (denoted on the left). f, Gene expression for genes associated with 5 groups of CPs as in e but shown as box plots. n = 646, 363, 1842, 1885 and 179 CPs, for Groups 1 to 5, respectively. (d and f) boxes: median and interquartile range; whiskers: 5th and 95th percentiles. g, Example of differentially activated alternative promoters, h, Example of differentially activated closely-spaced TSSs (g and h: merge of three independent biological replicates).