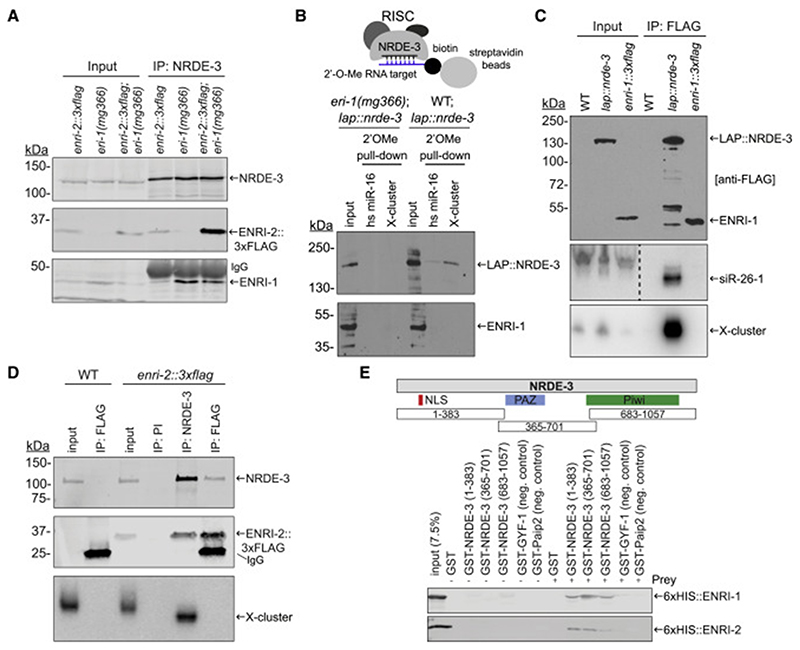
Figure 3. ENRI-1 and ENRI-2 interact with unloaded NRDE-3.
(A) Total lysates and NRDE-3 IPs from enri-2::3xflag, eri-1(mg366), and enri-2::3xflag;eri-1(mg366) embryos were probed with NRDE-3, FLAG and ENRI-1 antibodies. (B) Top: Schematic of the 2’O-methyl pull-down. Bottom: Input and pull-down fractions were probed with FLAG and ENRI-1 antibodies. (C) RNA extracted from FLAG IPs performed in transgenic lap::nrde-3 and enri-1::3xflag embryos was probed for 22G-RNAs mapping to siR-26-1 and X-cluster. FLAG antibody was used to detect both LAP::NRDE-3 and ENRI-1::3xFLAG. Dashed line indicates an unrelated lane was removed from the siR-26-1 northern blot. (D) RNA extracted from NRDE-3 and ENRI-2::3xFLAG IPs was probed for the 22G-RNAs mapping to the X-cluster. PI = pre-immune serum. NRDE-3 antibody and FLAG antibody were used to detect NRDE-3 and ENRI-2::3xFLAG respectively. (E) Top: schematic outlining the fragments of NRDE-3 fused to GST. Bottom: 50% of the pull-down was loaded onto a 10% gel for western blot analysis using an anti-6xHIS antibody. The other 50% was loaded onto a gel for Coomassie staining (Figure S3E, F). Each pull-down was performed at least 4 times and a representative blot for each is shown. See also Figure S3.