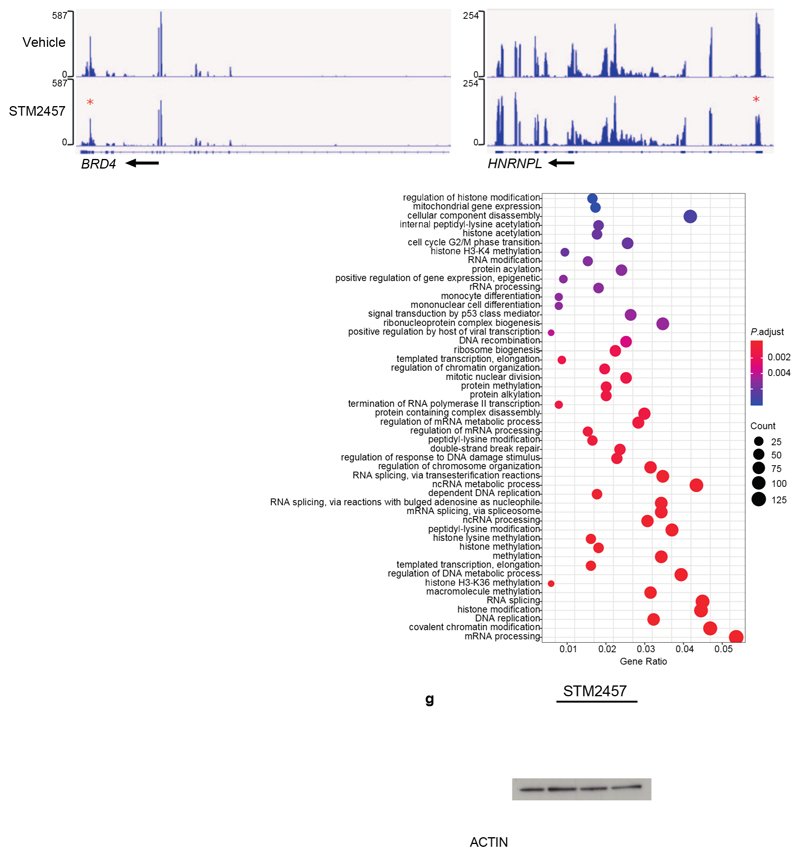
Extended Data Figure. 5. Pharmacological inhibition of METTL3 significantly reduces m6A on leukaemia-associated substrates.
a) Overlap between METTL3-dependent m6A poly-A+ RNAs in MOLM-13 cells either treated with 1 µM STM2457 or with genetic downregulation of METTL3 from Barbieri et al7. b) Overlap between differential downregulated m6A peaks in MOLM-13 cells either treated with 1 µM STM2457 or with genetic downregulation of METTL3 from Barbieri et al7. c) Genomic visualization of the m6A-meRIP normalized signal in MOLM13 cells following treatment with vehicle (DMSO) or 1 µM STM2457 for the METTL3-dependent m6A substrates BRD4 and HNRNPL (red stars indicate loss of m6A signal). d) m6A-meRIP-qPCR analysis of METTL3-dependent and METTL3-independent m6A substrates normalized to input in MOLM-13 cells treated for 24 or 48 hours with either vehicle (DMSO) or 1 µM STM2457 (mean ± s.d., n=3). e) Gene ontology analysis of differentially m6A-methylated mRNAs upon treatment with 1 µM STM2457. f) RT-qPCR quantification of METTL3 and DICER1 in total RNA samples from MOLM-13 cells treated with vehicle or STM2457 (mean +/- s.d., n=3). g) Western blot for METTL3, METTL14, DDX3X, DICER1 and ACTIN in MOLM-13 cells treated with 10, 5 and 1 µM of STM2457 or vehicle (DMSO) for 72 hours (n=3). two-tailed Student’s t-test; n.s., not significant; KD, knockdown.