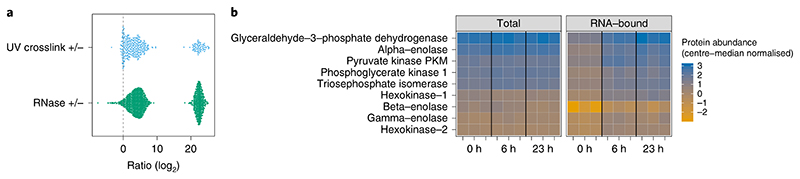
Fig. 7. OOPS MS results.
a, Expected results for SILAC cross-link (CL) +/− and RNase +/− experiments using U2OS cells. Proteins with no detectable signal in the negative-control sample are given a pseudo-value equivalent to the average intensity in the CL or RNase samples, respectively. These pseudo-values represent proteins highly enriched in the CL or RNase conditions. log ratios close to zero represent proteins without clear enrichment, which might be considered to be non-RNA-binding. Adapted from ref. 5, n = 4. b, Visualization of parallel total and RNA-bound protein abundances in a nocodazole arrest (0 h) and release (6 h and 23 h) experiment. Protein abundances are center-median normalized within each sample and are thus relative to the total protein abundance in a given sample and only comparable within total and RNA-bound quantifications, not between them. Shown here are the members of the glycolysis pathway. A clear increase in RNA-bound protein abundance between arrest and release time points is observed for many pathway members without any appreciable change in total protein abundance. n = 3 for each time point. Data presented are taken from ref. 5 and made available alongside plotting code in an R markdown notebook (see Supplementary Data Set 1).