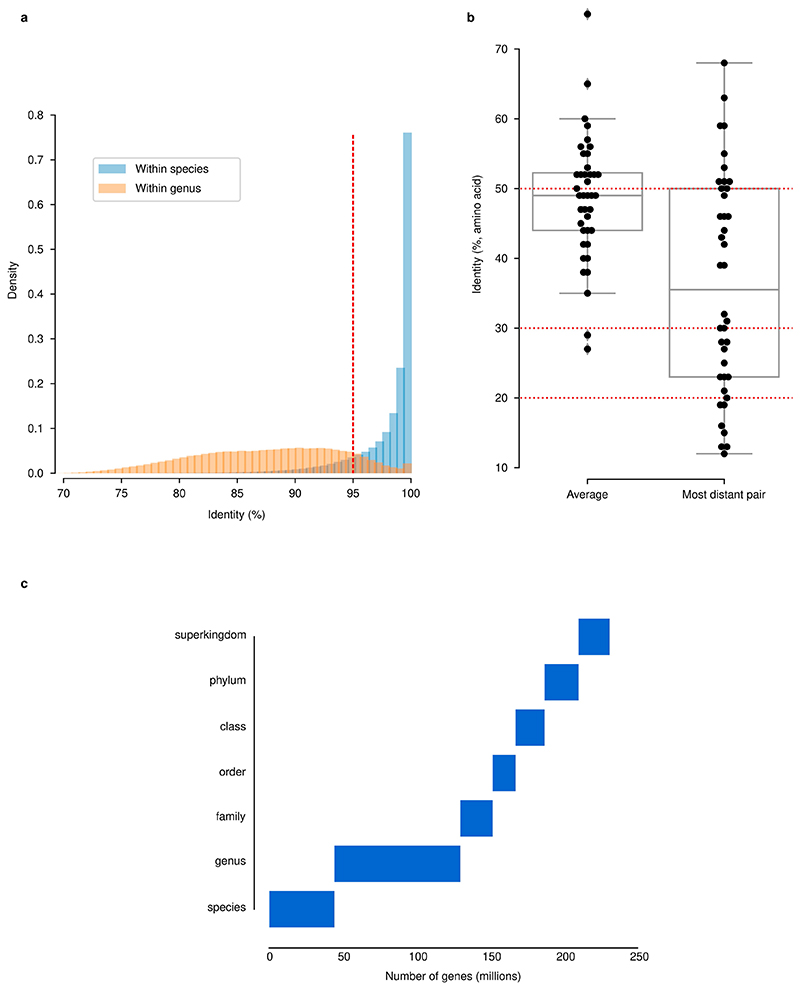
Extended Data Fig. 2. Identity thresholds and their relationship to taxonomy and function in the GMGCv1.
(a) A 95% nucleotide identity threshold is a proxy for species. Shown is nucleotide identity of closest gene homolog within the same species or within the same genus (excluding within-species comparisons). The threshold used in this work (95%) is marked with a dashed red line. (b) Within well-conserved, universal, 40 single-copy orthologues (see Methods), the average pairwise amino acid identity is 49%, albeit with a wide range (27-75%) when considering within-orthologue averages. In dashed red, the thresholds used for building protein families are highlighted. Boxplots display quartiles and ranges (see Methods). (c) Proportion of genes annotated at each taxonomic level.