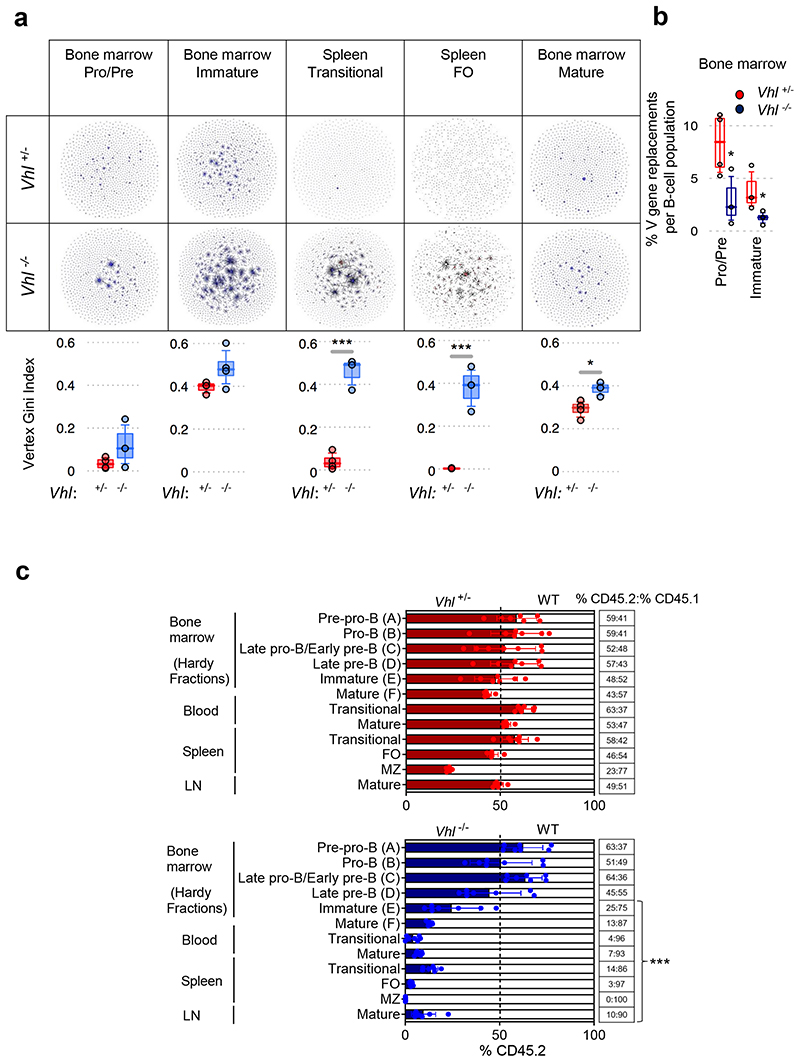
Figure 3. Regulated HIF-1α activity is required for the generation and diversification of the pre-immune B cell repertoire.
(a) Clonal analysis of IgHV-D-J DNA rearrangements from BM B cells and IgHV-D-J RNA rearrangements from spleen B cells. Table contains sequencing-networks demonstrating that in Vhl-/-Mb1-cre B cells, IgHV diversity is reduced (number of dots) and the maximum clone size (relative size of dots) is increased. Below, quantification using the Vertex Gini Index. (b) High-throughput detection of IgHV secondary rearrangements in Vhl+/-Mb1-cre and Vhl-/-Mb1-cre B cells from BM (DNA). Plotted, frequency of unique BCR-sequences with the same stem region. (a-b) Gated on B cells Pro-B, Pre-B B220+IgM-, immature and transitional B220+IgM+IgD-, BM mature B220+IgM+IgD+ and FO B220+IgM+IgD+CD23+CD21+,*P<0.05,***P<0.005 one-sided Wilcoxon-tests. Graphs; box lines show the 25th, 50th and 75th percentiles; whiskers show the 10th and 90th percentiles. n=4 biologically independent mice per genotype from one experiment. (c) The relative proportion of B cell subsets in lethally-irradiated CD45.1 mice reconstituted for 8 weeks with 1:1 mixtures of CD45.1 WT and CD45.2 Vhl+/-Mb1-cre or Vhl-/-Mb1-cre BM. Gated on Pre-pro-B (FrA:B220+CD43+CD24-/lowBP-1-), Pro-B (FrB:B220+CD43+CD24+BP-1-), Late pro-B/Early pre-B (FrC:B220+CD43+/lowCD24+BP-1+), Late pre-B (FrD:B220+CD43-IgM-IgD-), immature (FrE:B220+CD43- IgM+IgD-), BM mature (FrF B220+CD43-IgM+IgD+), blood transitional (B220+IgM+IgD-), blood and ILN mature (B220+IgM+IgD+), spleen transitional (B220+CD93+), FO and MZ (gated as in Figure 1). Filled columns; mean %CD45.2, clear columns; mean %CD45.1, error bars S.D.; comparison between Vhl+/-Mb1-cre and Vhl-/-Mb1-cre B cells by two-way ANOVA Bonferroni post-test ***P<0.001, n=7 biologically independent mice per genotype; data, representative of two independent experiments. (a-c) Symbols represent individual mice.