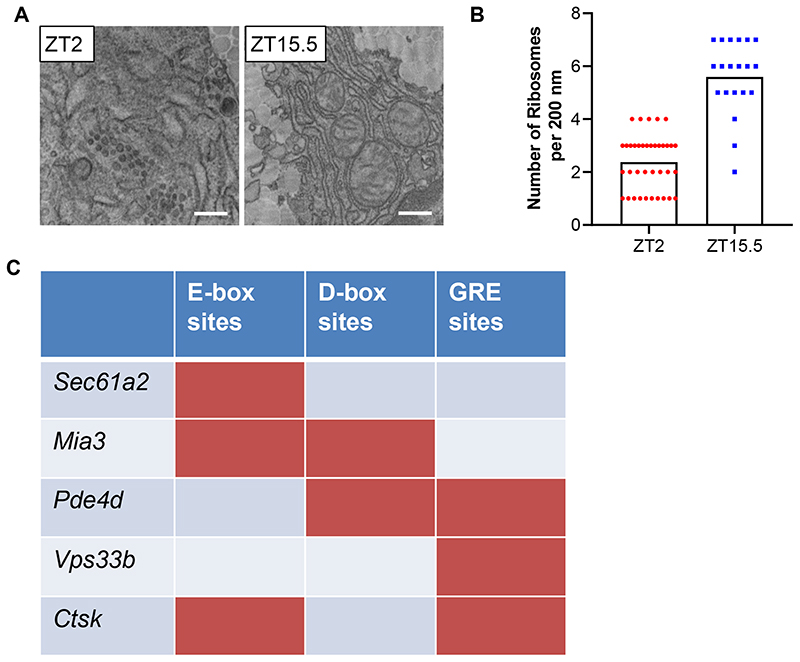
Extended Data Fig. 3. Rhythmic ribosome docking.
a, Images of electron microscopy of transverse sections across mouse tendons fibroblasts in vivo showing the endoplasmic reticulum (ER) at different time points, n = 2 biological repeats. Bars 500 nm. b, Quantification of ribosome docking onto ER show differences at ZT2 and ZT15.5 in tendon fibroblasts in vivo, n = 2 biological repeats with 37 (ZT2) and 20 ZT15.5 regions scored across the two samples. Samples were scored by two independent researchers, all samples were blinded (p = 0.00000000034, two-tailed unpaired t-test). The mean and SD for each sample is shown. c, The transcription factor binding site prediction tool Contra v3 http://bioit2.irc.ugent.be/contra/v3 was used to search for binding sites in the promoter and upstream sequences of the genes shown. The default (pragmatic)positional weight matrices (PWMs) score matching stringency in a 500 base promoter region was used. E-box: Ebox, CLOCK/BMAL, Myc, Myc/Max. Cry1/Cry2 binding sites are E- boxes/CLOCK/BMAL and ROR/REV-ERB. D-box: E4BP, NFIL3, ATF2, CEBP. GRE: GRE, NR3C1. ROR: RORA, RORB, RORC. See also Statistical Source Extended Data Fig. 3. Source data