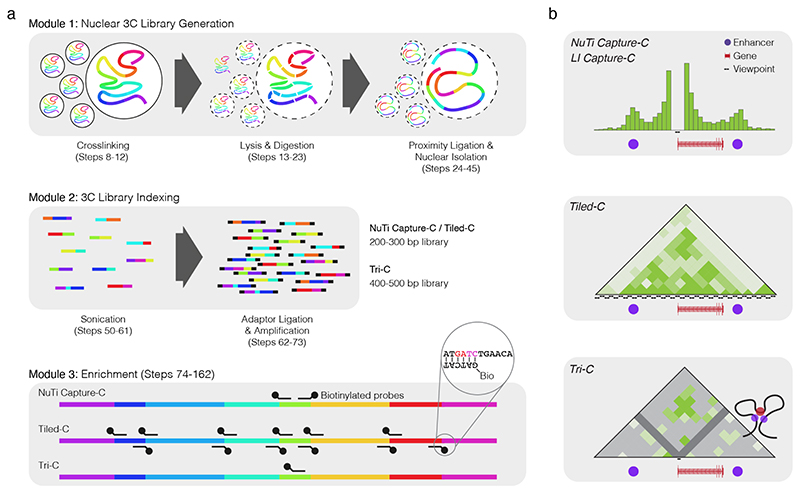
Figure 1. Capture-C is modular and adaptable for characterizing chromatin folding.
a, The Capture-C family of methods involves three distinct modules. In the first module a Nuclear 3C library is generated from 2% formaldehyde fixed cells that are lysed, then permeabilised with SDS, and digested with a frequent 4-base cutter (DpnII or NlaIII). Proximity ligation re-arranges the genome order to reflect spatial 3D organisation. Finally, for this module, centrifugation is used to separate DNA from ruptured nuclei from DNA in intact nuclei, which contains more informative 3C material. Library indexing in module 2 is performed using standard next-generation sequencing kits with sonication providing unique ends for PCR duplicate filtering. For Tri-C, gentler sonication is used to generate longer fragments which contain multiple ligation junctions. The third module is the most diverse, with a unique oligonucleotide design for each method. NuTi Capture-C uses a pair of oligonucleotides from the same strand of DNA that overlap restriction digestion sites of disperse fragments. For Tiled-C the same approach is used, however contiguous fragments are targeted and double stranded oligonucleotides have typically been used. In Tri-C a single oligonucleotide in the centre of a short restriction fragments enriches for sonication fragments with multiple ligation junctions. b, Schematic of results for a hypothetical locus, with one gene (red) and two enhancers (purple circles). NuTi Capture-C, or the low-cell variation LI Capture-C, from the promoter can be used to show direct interactions with both enhancers, Tiled-C produces a Hi-C like interaction map showing the three elements are in a TAD-like regulatory domain, and Tri-C shows that the two enhancers can be found simultaneously interacting with each other and the promoter at single alleles.