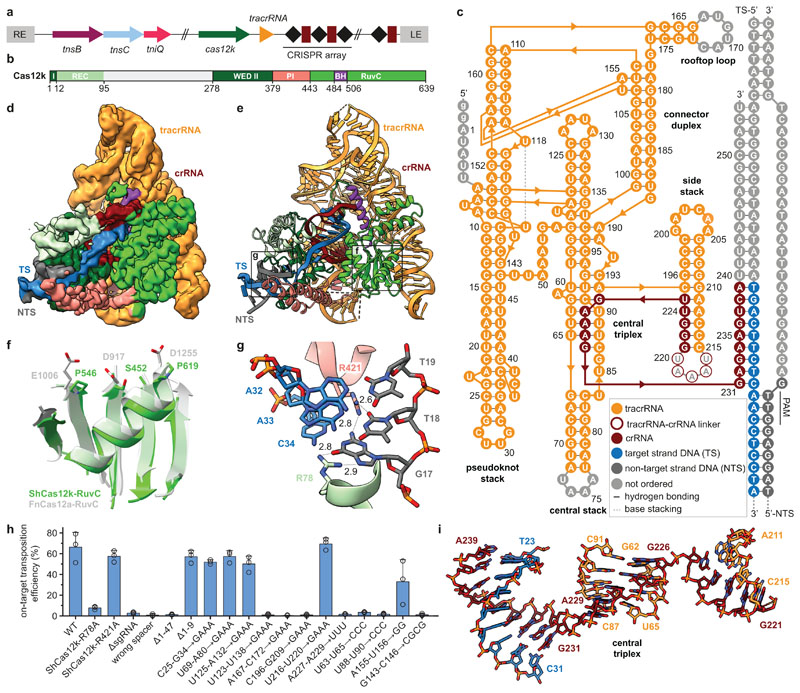
Fig. 1. Cryo-EM structure of the ShCas12k-sgRNA-target DNA complex.
a, Schematic of the type V-K CRISPR-associated transposon system from Scytonema hofmanni. LE, RE - left and right transposon ends. b, Domain organization of ShCas12k. REC, recognition lobe. WED, wedge domain. PI, PAM interacting domain. BH, bridging helix. c, Schematic of the sgRNA structure. d-e, Electron density map (d) and structural model (e) of the ShCas12k-sgRNA-target DNA complex. TS, target DNA strand; NTS, non-target DNA strand. f, Detail of the inactivated ShCas12k RuvC domain overlaid with the FnCas12a RuvC domain (PDB: 6i1k)30. g, Detailed view of PAM recognition. Hydrogen bonds are depicted with dotted lines. Distances are indicated in Å. h, Site-specific transposition activity in E. coli of ShCAST systems containing mutations in PAM interacting residues, as determined by droplet digital PCR (ddPCR) analysis. Data are presented as mean ± s.d. (n=3 biologically independent replicates).i, Details of the sgRNA central triplex.