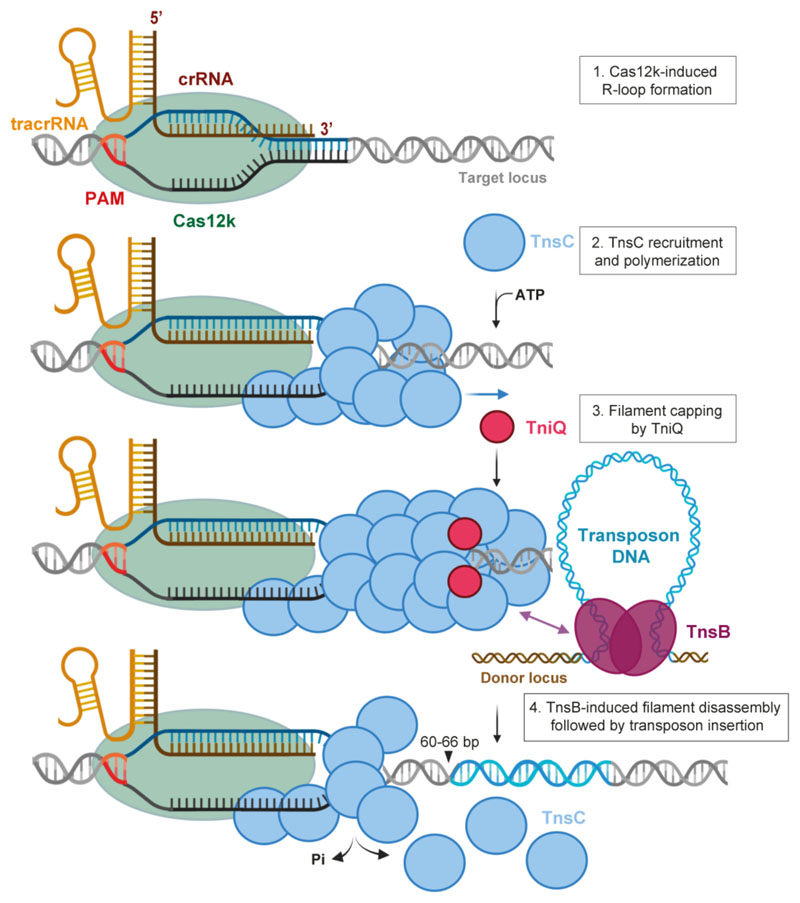
Extended Data Fig. 8. Mechanistic model of Cas12k-directed transposition.
Schematic diagram depicting transposition of type V-K CRISPR-associated transposons. Cas12k in association with a crRNA-tracrRNA dual guide RNA recognizes target DNA sequences, forming a partial R-loop structure. Full R-loop is formed upon recruitment of TnsC by interactions with DNA-bound Cas12k, which nucleates ATP-dependent formation of a helical filament around structurally remodeled DNA. Filament growth is restricted by TniQ capping the Cas12k-distal end of the TnsC filament. The TnsC filament serves as a recruitment platform for TnsB, which interacts directly with TnsC and stimulates its ATPase activity. This leads to filament disassembly, making DNA downstream of the PAM accessible for transposon insertion. In the post-hydrolysis state, TnsC forms a single-turn helical hexamer around target DNA, possibly acting as a molecular ruler to define the spacing between the Cas12k binding and transposon insertion sites (60-66 nt downstream of the PAM).