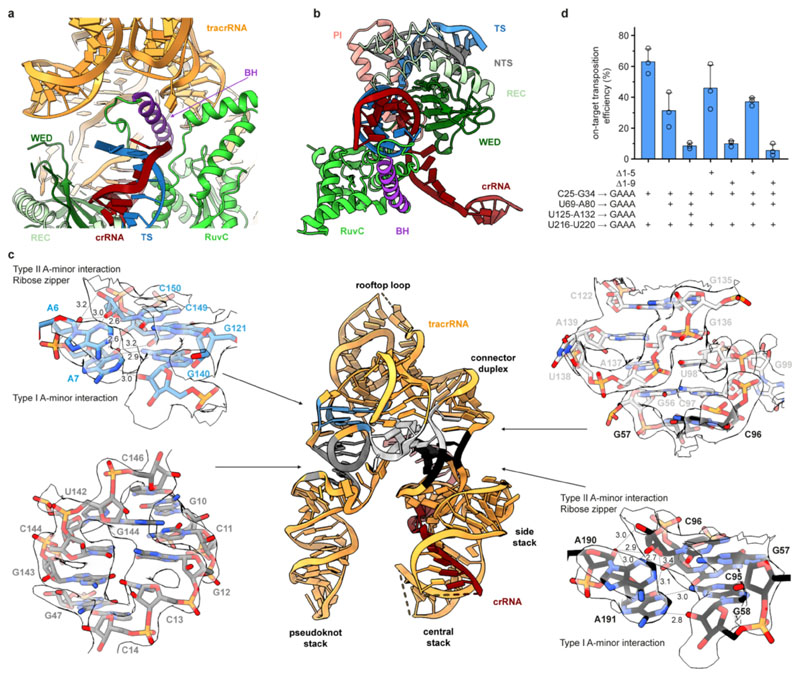
Extended Data Fig. 4. Structural features and functional analysis of ShCAST sgRNA.
a, Close-up view of the RNA:DNA duplex in proximity of the Bridge Helix (BH). b, Close-up view of the sgRNA-TS DNA heteroduplex in the ShCas12k-sgRNA-target DNA complex. c, Structural model of the ShCas12k sgRNA with detailed views of the 5’ terminal segment (top left, 6.6 σ contour level) and the triplex junction (bottom right, 9.0 σ contour level) of the tracrRNA forming ribose-zipper and A-minor interactions, the pseudoknot duplex (bottom left, 9.0 σ contour level) and the central stack junction (top right, 7.6 σ contour level). d, Droplet digital PCR (ddPCR)-based analysis of the transposition activity of structure-based sgRNA scaffold mutants in the ShCAST system. Data are presented as mean ± s.d. (n=3 biologically independent replicates).