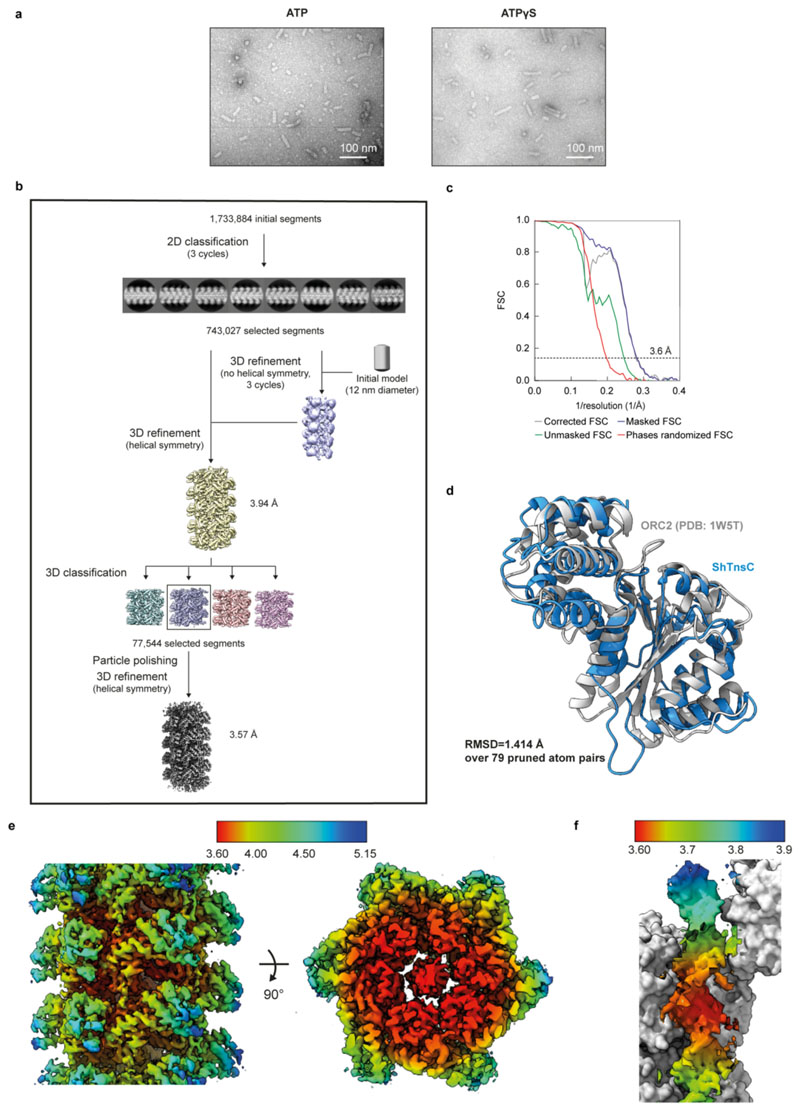
Extended Data Fig. 5. ShTnsC filament formation and cryo-EM image processing of ShTnsC•AMPPNP•dsDNA helical filaments.
a, Representative negative-stain EM micrographs of ShTnsC in the presence of a 92 bp dsDNA and ATP or ATPγS. Scale bars, 100 nm. Magnification, 120,000x. Experiment was repeated twice independently with similar results. b, Cryo-EM data processing workflow for the ShTnsC-dsDNA complex. Selected 2D class averages used in image reconstruction and intermediate and final reconstructions are shown. c, Fourier Shell Correlations (FSC) of ShTnsC filament reconstruction from two independently refined half-maps. The gold-standard cutoff (FSC = 0.143) is marked with dashed lines. The resolution value of the FSC corrected curve at this level is indicated. d, Structural alignment of the cryo-EM structure of ShTnsC (blue) and the crystal structure of the Aeropyrum pernix ORC2 protein (grey; PDB ID: 1W5T52) used as initial homology model for model building (single protomers). The root mean square deviation (RMSD) between the structures is 1.41 Å over 79 pruned atom pairs, as calculated in Chimera. e, Local resolution estimation (Å) for the cryo-EM volumes of the ShTnsC•AMPPNP•dsDNA helical filaments. f, Local resolution estimation (Å) of the bound DNA duplex.