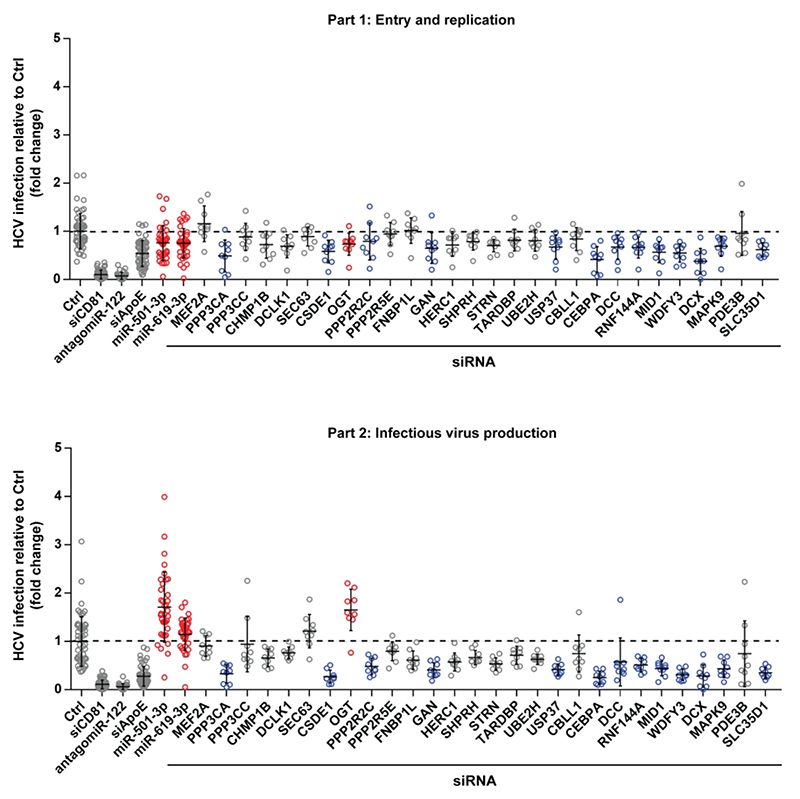
Figure 2. OGT is a novel host cell factor involved in the late steps of the HCV life cycle.
Huh7.5.1 cells were transfected with a set of siRNAs against 28 predicted targets of miR-501-3p and/or miR-619-3p, and infected with HCVcc JcR2A according to the two-step protocol depicted in Fig. 1A. siCD81, antagomiR-122 and siApoE were used as loss-of-function controls to perturb HCV entry, translation/replication and assembly, respectively. miR-501-3p and miR-619-3p, which were ineffective in part 1 of the screen but enhanced HCV infection in part 2, were transfected in parallel. HCV infection was quantified as fold change of luciferase activity with respect to negative control (siCtrl). Results for different replicates are shown as individual points. For each gene, median fold change of luciferase activity ± s.d. is shown as black horizontal lines. The dashed line indicates a fold change of 1. Data are from three independent experiments in triplicate. Results for miR-501-3p, miR-619-3p and siOGT that increase HCV infection in part 2 are depicted in red. Results for siRNA targeting PPP3CA, CEBPA, MID1, WDFY3, DCX, SLC35D1, CSDE1, GAN, USP37, MAPK9, DCC, RNF144A, or PPP2R2C that significantly modulated HCV infection in part 1 and/or part 2 but did not phenocopy the effect of miR-501-3p and miR-619-3p are depicted in blue.