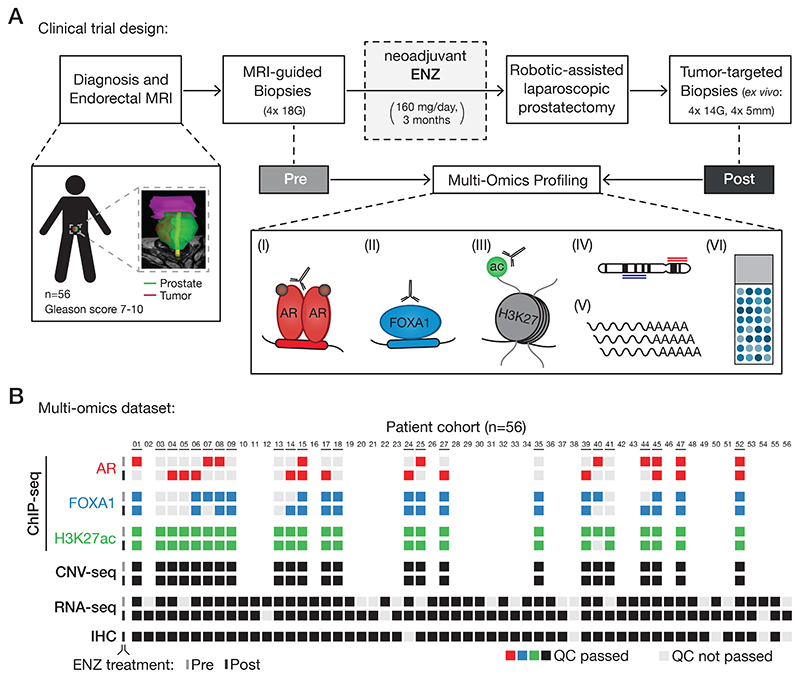
Figure 1. Clinical trial design and omics data sample collection.
(A) Study design of the DARANA trial (NCT03297385). Multi-omics profiling, consisting of (I) Androgen Receptor (AR) ChIP-seq, (II) FOXA1 ChIP-seq, (III) H3K27ac ChIP-seq, (IV) DNA copy number sequencing (CNV-seq), (V) gene expression profiling (RNA-seq) and (VI) immunohistochemistry (IHC) analysis, was performed on MRI-guided biopsy samples prior to ENZ treatment (Pre) and tumor-target prostatectomy specimens after 3 months of neoadjuvant ENZ therapy (Post).
(B) Overview of data availability and quality control analyses for each sample. Individual data streams are indicated separately with ChIP-seq for AR (red), FOXA1 (blue), H3K27ac (green), CNV-seq, RNA-seq and IHC (all black). The ENZ treatment status indicates the pre-treatment (top) and post-treatment samples (bottom) per omics dataset. Samples not passing QC (light gray) were successfully applied for focused raw data analyses. Blank spots for ChIP-seq or CNV-seq samples indicate that the fresh-frozen material didn’t pass the tumor cell percentage cutoff of ≥ 50%.