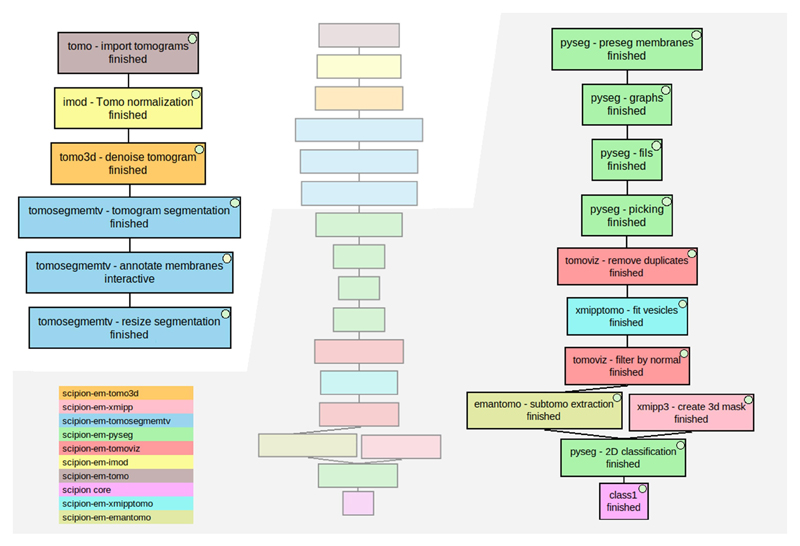
Fig. 9.
Directional picking workflow to pick membrane-associated ribosomes of an in situ tomogram (EMD-10439). The workflow in the middle represents the whole workflow, while the images on both sides are zoomed-in parts of it to provide better visualization. Different colors were used to represent each plugin involved. The color legend is on the bottom left. The different stages described in the text are (i) boxes named “tomo - import tomograms” (in brown), “Imod - Tomo normalization” (in yellow), and “tomo3d - denoise tomogram” (in orange), (ii) the 3 blue boxes named “tomosegmemtv - tomogram segmentation”, “tomosegmemtv - annotate membranes” and “tomosegmemtv - resize segmentation”, (iii) the green boxes named “pyseg - preseg membranes”, “pyseg - graphs”, “pyseg - fils” and “pyseg - picking”, (iv) boxes named “tomoviz - remove duplicates” (in red), “xmipptomo - fit vesicles” (in light blue) and “tomoviz - filter by normal” (in red), and (v) boxes named “emantomo - subtomo extraction” (in dark yellow), “xmipp3 - create 3d mask” (in light red), “pyseg — 2D classification” (in green) and “class1” (in pink).