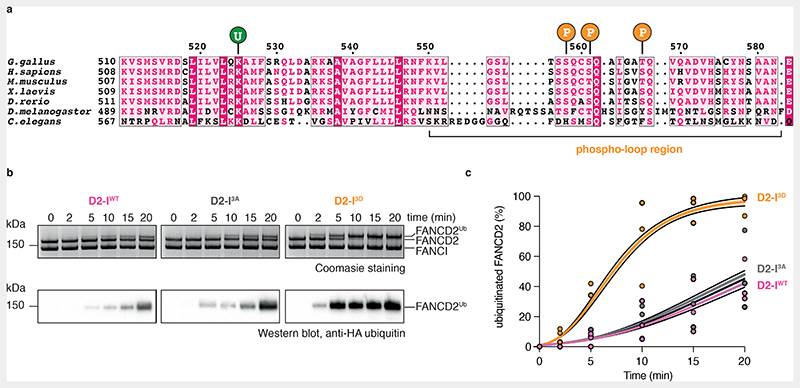
Fig. 1. Phosphomimetic mutations of conserved ATR phosphorylation sites in the FANCI phospho-loop stimulate monoubiquitination of FANCD2.
(a) Multiple sequence alignment of residues 510-583 from G. gallus FANCI and the corresponding region in other FANCI orthologues. Highly-conserved residues are shaded in magenta; partially-conserved residues are shaded in light pink. The FANCI-K525 ubiquitination site is marked with a ‘U’ and each of the three phosphorylation sites within the conserved S/TQ motifs are marked with a ‘P’. The phospho-loop is indicated. (b) Monoubiquitination assays of wild-type (WT), phosphodead (3A) and phosphomimetic (3D) D2-I complexes. The level of FANCD2 ubiquitination was analyzed using SDS-PAGE and Coomassie staining (top) and by western blotting with an anti-HA antibody against HA-tagged ubiquitin (bottom). Ubiquitination is visible as an increase in apparent molecular weight of FANCD2. These data are representative of experiments performed independently three times. (c) Quantification of monoubiquitination assays from panel b. Data obtained from three independent assays were quantified as percentage of ubiquitinated FANCD2. Replicate data were fitted using a global non-linear regression function as described in the Methods. Data points of each replicate (symbols), the corresponding mean (central solid line) and the 95% confidence interval (shaded area between dashed lines) are shown.