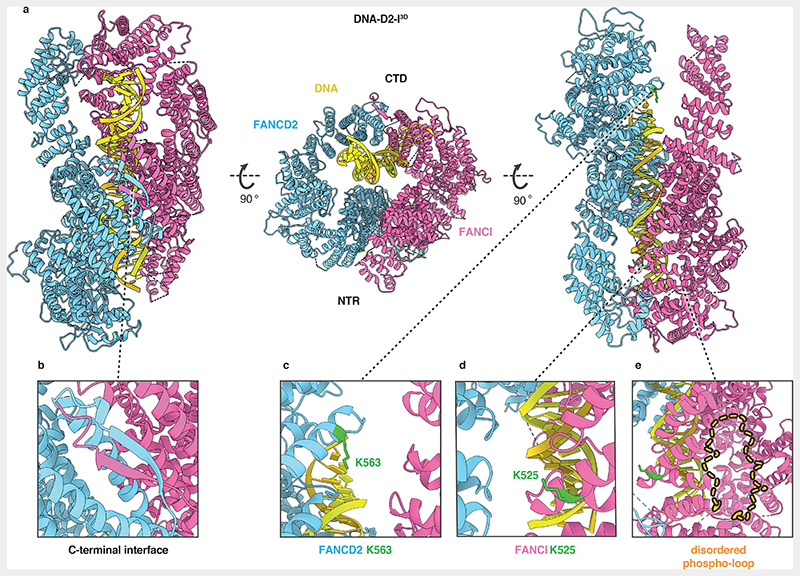
Fig. 3. Closed, DNA-bound D2-I3D is poised for monoubiquitination.
(a) Cartoon representation of the DNA-bound D2-I3D atomic model showing a front view over the C-terminal interface (left), an end-on view down the DNA channel with the N-terminal region (NTR) and the C-terminal domain (CTD) both visible (middle) and a back view of the N-terminal dimerization interface where the ubiquitination sites are located (right). (b) Close-up view of the C-terminal dimerization interface that forms upon D2-I3D closure around DNA, showing the zipper-like mechanism previously observed in structures of the ubiquitinated or the FA core complex- and DNA-bound wild-type D2-I. (c-d) Close-up of FANCD2-K563 (c) and FANCI-K525 (d) represented as sticks (green) show that these target lysines are solvent-accessible and not occluded in the N-terminal dimerization interface. (e) The phospho-loop is flexible, and could not be modelled. A close-up view of this region with an illustration of a fully extended phospho-loop (dashed orange line) shows its potential position in the context of a closed DNA-bound D2-I3D complex.