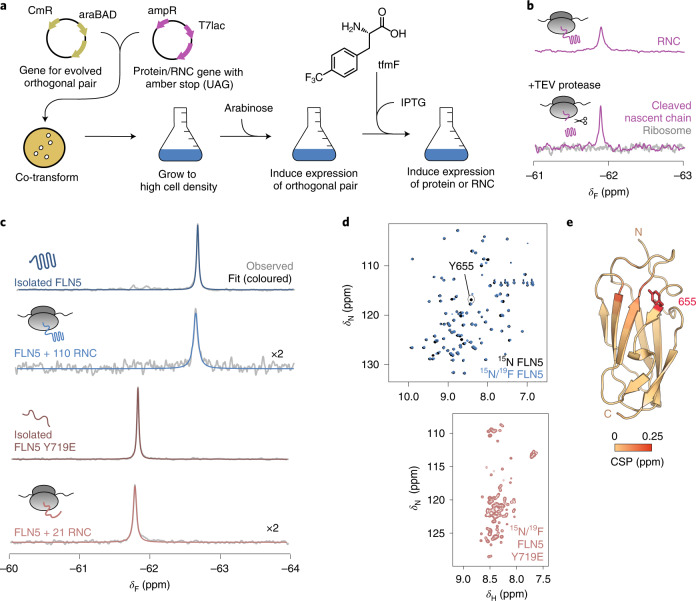
Fig. 1. Site-specifically 19F-labelled RNCs report on the folding of FLN5 on and off the ribosome.
a, Schematic of production of 19F-labelled RNCs (Methods). CmR, cloramphenicol resistance gene; araBAD, l-arabinose operon; ampR, ampicillin resistance gene; T7lac, T7 promoter inducible by isopropyl ß-d-1-thiogalactopyranoside (IPTG). b, The 19F NMR spectra of a RNC with a cleavable FLN5 domain, before and after addition of tobacco etch virus (TEV) protease and purification of component parts (Extended Data Fig. 1). c, The 19F NMR spectra of isolated FLN5 and FLN5 + 110 RNC, and isolated FLN5 Y719E and FLN5 + 21 RNC. Observed and fitted spectra are shown in grey and red/blue respectively (298 K, 500 MHz). δF, 9F chemical shift. RNC spectra magnified by a factor of ×2. d, The 2D 1H,15N NMR (selective optimized flip angle short transient (SOFAST) heteronuclear multiple quantum coherence (HMQC)) spectra of 15N-labelled and 15N/19F-labelled isolated FLN5 and FLN5 Y719E (298 K and 283 K, respectively; 800 MHz). δN, 15N chemical shift; δH, 1H chemical shift. e, Crystal structure of FLN5 (Protein Data Bank (PDB) no. 1QFH) coloured by residue-specific 1H,15N amide backbone chemical shift perturbations (CSP) observed following 19F incorporation at position 655 (Extended Data Fig. 3). The N and C termini are shown.