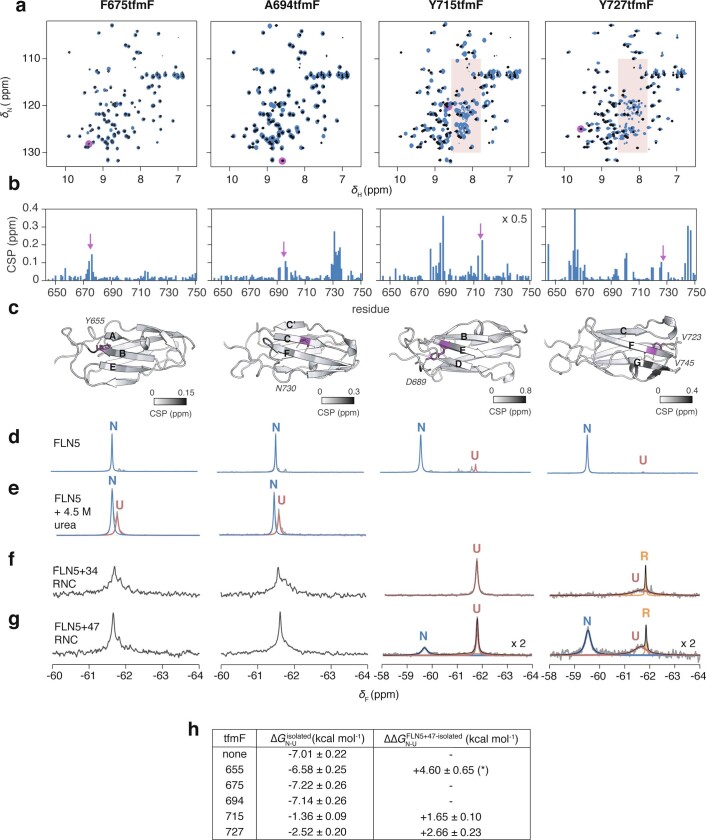
Extended Data Fig. 6. 19F NMR spectroscopy of FLN5 with tfmF incorporation at positions F675, A694, Y715, and Y727.
a, 2D 1H,15N-SOFAST HMQC spectra of FLN5 without (black) and with tfmF-incorporation (blue). Resonance corresponding to incorporation site is absent in each tfmF-labelled FLN5 construct, as highlighted in magenta. Red shading indicates disordered resonances resulting from destabilisation by tfmF-incorporation in solvent-inaccessible positions (see h). b, 1H,15N-correlated chemical shift perturbations measured from spectra shown in a upon tfmF-incorporation. c, Location of tfmF-incorporation (magenta) on the crystal structure of FLN5 (1qfh), coloured according to chemical shift perturbations. Contacts made by non-fluorinated FLN5 at the label site are shown by dashed lines and the contacted residues labelled. d, 1D 19F NMR spectra of tfmF-incorporated FLN5. Arrows indicate the appearance of a disordered resonance, consistent with 1H,15N-correlated NMR observations. e, 19F NMR spectra of tfmF-incorporated FLN5 incubated in 4.5 M urea, used to determine the ∆∆G of tfmF-incorporation by comparison with 19F NMR spectra of FLN5 labelled at position 655 and incubated in 4.5 M urea, as shown in Extended Data Fig. 5. f, 19F NMR spectra of tfmF-incorporated FLN5 + 34 RNC. g, 19F NMR spectra of tfmF-incorporated FLN5 + 47 RNC. Ribosome-released species are indicated by orange arrows. For spectra with well-resolved resonances, the data were fitted to Lorentzian line shapes. The broad linewidth of the unfolded state for tfmF727 FLN5 + 47 is consistent with its position in the ribosome-interacting segment of the domain17, and is reduced by ~25% relative to its linewidth in FLN5 + 34. The spectra of RNCs tfmF-labelled at positions 675 and 694 show highly overlapped resonances and so we were unable to accurately fit the peaks. h, Gibb’s free energies of tfmF-incorporated isolated FLN5, determined by quantification of native state peak integrals of spectra shown in d and e, and free energy differences (∆∆GN-U) between the ribosome-bound (with 47-residue linker) and isolated native states. The ∆∆GN-U for tfmF655-labelled FLN5 (labelled *) is estimated based on a population of U determined from the spectral noise. 19F-labelling at positions 715 and 727 show reduced destabilisation of N on the ribosome relative to when labelled at position 655 for FLN5 + 47 RNC (similar results are obtained when including I1 and/or I2 states); tfmF side chains in positions 715 and 727 therefore form native-like tertiary contacts before those in 655 are formed in the intermediate states, consistent with a folded core comprising the B-F strands.