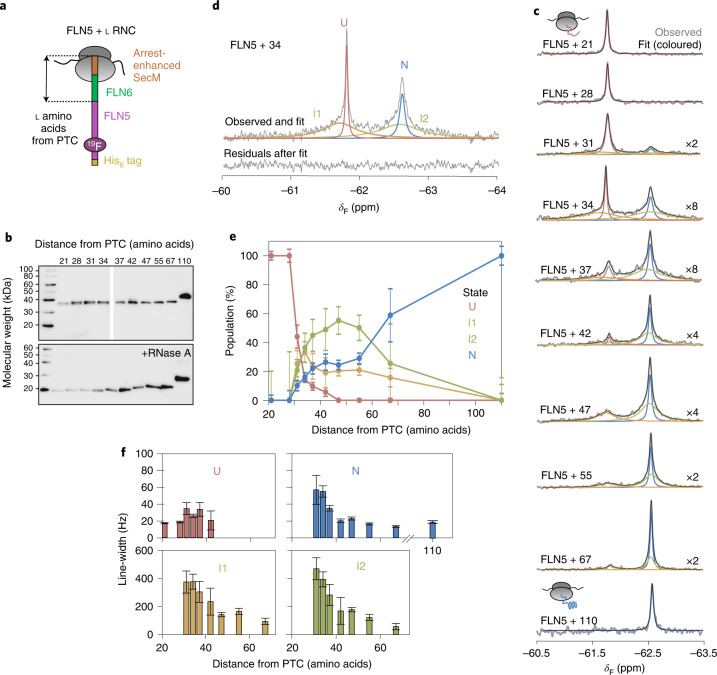
Fig. 2. Co-translational folding of FLN5 monitored by 19F NMR spectroscopy.
a, Design of FLN5 RNCs in which FLN5 is tethered to the PTC via a linker sequence comprising a variable number of FLN6 residues and an arrest-enhanced SecM stalling motif. b, Anti-hexahistidine western blot of purified FLN5 RNCs, with and without ribonuclease A (RNase A) treatment. Representative data shown from two independent repeats. c, The 19F NMR spectra of FLN5 RNCs with increasing distance from the PTC. Observed spectra shown in grey were fitted and peaks assigned to U, I1, I2 or N states (coloured), with the sum of the fits shown in black. NMR data were multiplied with an exponential window function (10 Hz line broadening factor) before Fourier transformation. d, The 19F NMR spectrum of FLN5 + 34 RNC, processed with a line broadening factor of 5 Hz. Residual spectrum after fitting is shown below. e, Folding of FLN5 on the ribosome, measured using 19F NMR line-shape fits. f, Line-widths measured by line-shape fits of spectra as shown in c. All error bars indicate errors calculated by bootstrapping of residuals from NMR line-shape fittings.