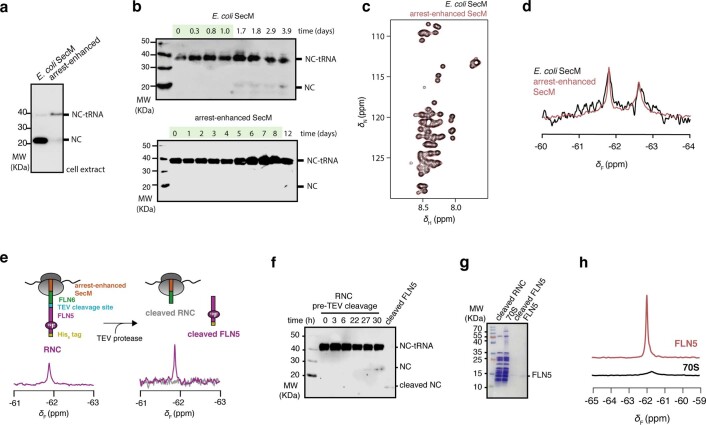
Extended Data Fig. 1. Development of in vivo site-selective 19F-labelling of arrest-enhanced RNCs using amber suppression.
a, Anti-histidine western blot of cell extracts following expression without further purification of FLN5 + 31 RNC translationally stalled using SecM deriving from E. coli, and an arrest-enhanced variant of SecM based on the sequence deriving from Mannheimia succiniciproducens66 with the sequence ‘FSTPVWIWWWPRIRGPP’. A higher amount of released nascent chain relative to ribosome-bound (that is tRNA-bound) nascent chain (NC-tRNA) is interpreted as higher ribosome turnover/read-through, and thus weaker translation arrest. b, Anti-histidine western blot of samples of purified FLN5 + 31 A3A3 RNC17 with E. coli SecM (upper) and arrest-enhanced SecM (lower) incubated at 10˚C. Green shading indicates time during which exclusively ribosome-bound (tRNA-bound) nascent chain is detected. c, 2D 1H,15N-SOFAST HMQC spectra of a non-ribosome interacting FLN5 + 31 RNC variant with E. coli SecM (black) and arrest-enhanced SecM (red); no discernible difference was found. d, 1D 19F NMR spectra of FLN5 + 34 RNC with E. coli SecM (black) and arrest-enhanced SecM (red). No discernible difference was found, notwithstanding the significantly higher effective signal-to-noise provided by longer available data acquisition of the arrest-enhanced RNC. e, (left) 1D 19F spectrum of FLN5, 19F-labelled at position 691 and translationally stalled by the arrest-enhanced SecM motif, linked together with a linker comprising FLN6 residues and a TEV protease cleavage site. (right) 1D 19F spectrum following cleavage by TEV protease and purification of the two component parts to produce the cleaved RNC and cleaved FLN5. f, Anti-histidine western blot of RNC sample during NMR data acquisition and following TEV protease cleavage. g, Coomassie-stained SDS-PAGE of purified samples. h, 1D 19F spectra of purified (upper) FLN5, and (lower) 70S ribosomes purified from E. coli transformed with the plasmid encoding the orthogonal pair and exclusively grown in cultures supplemented with tfmF to achieve 100% tfmF labelling. Spectra are normalised to molar concentrations and number of experimental scans. These data demonstrate that even with 100% background labelling of the ribosome, its signal intensity remains substantially lower than that of FLN5. Western blots and gels show representative data from two independent repeats.