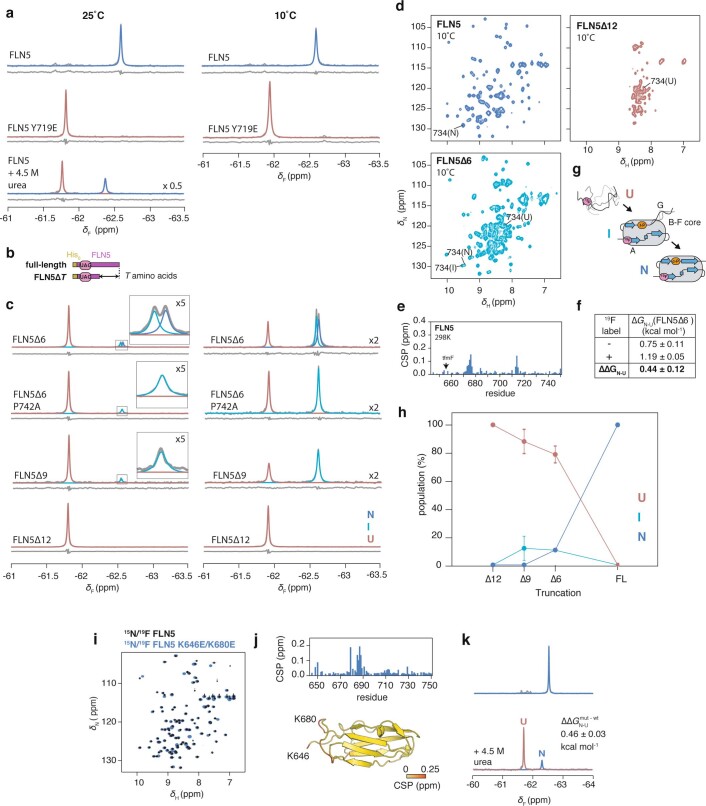
Extended Data Fig. 3. NMR spectroscopy of isolated variants of 19F-labelled FLN5.
a, 1D 19F NMR spectra of isolated FLN5, FLN5 Y719E and FLN5 in the presence of 4.5 M urea, recorded at 25˚C and 10˚C. No intermediate state population (>5%) was detectable under denaturing conditions. b, Design of C-terminal truncations of FLN5. c, 1D 19F NMR spectra of FLN5∆6, FLN5∆6 P742A, FLN5∆9, and FLN5∆12, recorded at 25˚C and 10˚C. Observed spectra (in grey) were fitted to Lorentzian line shapes and assigned to the various isolated states: natively folded (N, blue), intermediate (I, cyan), and unfolded (U, red). Residuals after fitting are shown below each spectrum. The P742A mutation results in destabilisation of the ∆6 N state, enabling us to attribute the intermediate state as having a cisP742 conformation30. d, 2D 1H,15N-SOFAST HMQC spectra of FLN5, FLN5∆6, and FLN5∆12, recorded at 10˚C. Assignment of residue R734 is shown as an example of resonances in N, I, and U states. e, 1H,15N SOFAST-HMQC chemical shift perturbations of isolated FLN5 following introduction of Y655tfmF, at 298 K. f, Incorporation of tfmF results in a small destabilisation in the natively folded state, as determined by integration of FLN5∆6 peak shown in c, and compared against previous measurements of non-fluorinated protein30. Errors propagated from bootstrapping of residuals from NMR line shape fittings. g, Schematic summarising length-dependent folding pathway of isolated FLN530. h, Length-dependent folding of isolated, 19F-labelled FLN5, determined by integration of spectra shown in a and d. Error bars indicate errors propagated from bootstrapping of residuals from NMR line shape fittings.i, 1H,15N-correlated NMR spectra of isolated 15N/19F-labelled FLN5 and FLN5 K646E/K680E. j, 1H,15N- chemical shift perturbations of FLN5 upon introduction of K646E/K680E mutations from analysis of spectra shown in a shown per residue in the plot, and coloured onto the FLN5 crystal structure below. k, 19F NMR spectra of FLN5 K646E/K680E in 0 and 4.5 M urea. The folded/unfolded populations of the latter were determined by lineshape fitting, and compared against those obtained for wild-type FLN5 (shown in a) to obtain its change in thermodynamic stability.